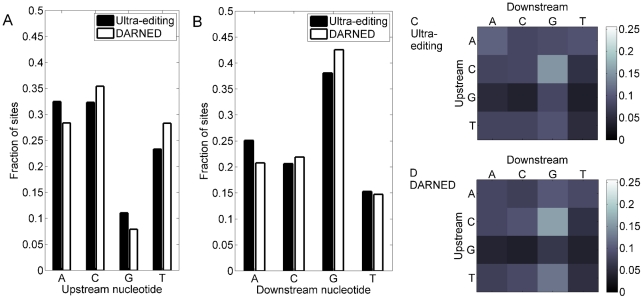
Figure 5. Sequence context of ultra-editing.
(A) The composition of (genomic) nucleotides upstream of the editing sites. Solid bars- ultra-editing sites; hollow bars- all previously known editing sites (from DARNED, the database of RNA editing [30]). Shown is the fraction of sites with each type of nucleotide. (B) Same as (a), for the nucleotide downstream of the editing site. The main editing motif for both DARNED and ultra-editing is a deficit in G upstream and an excess of G downstream of the editing site. (C) The fraction of each dinucleotide combination (upstream-downstream) for the ultra-editing sites. Brighter squares correspond to more frequent dinucleotides (color coded on the right). (D) Same as (c) for DARNED.