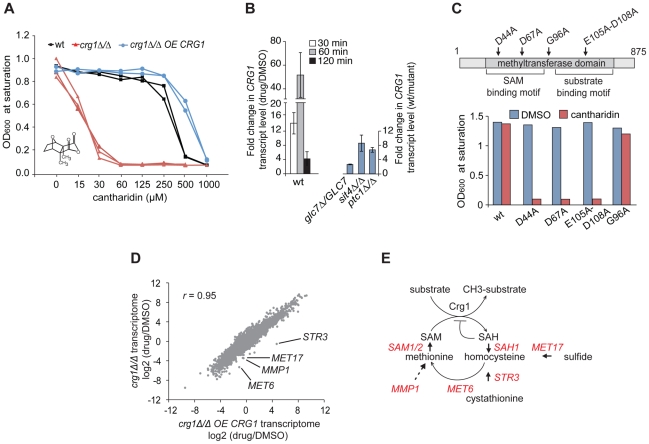
Figure 1. Functional SAM-dependent methyltransferase Crg1 is required for cantharidin response.
(A) CRG1 gene dose is important for cantharidin tolerance. Wt, crg1Δ/Δ and CRG1-overexpressing crg1Δ/Δ mutants were assessed in the presence of cantharidin in YPD. Dose-response curves were obtained by plotting OD600 at saturation point vs. tested drug concentrations. (B) Chemical (left panel) and genetic inhibition (right panel) of protein phosphatases result in the induction of CRG1. Wt, glc7Δ/GLC7 heterozygous, sit4Δ/Δ and ptc1Δ/Δ homozygous deletion mutants grown to mid-exponential phase were incubated with or without cantharidin (250 µM). For each time point, total RNA was extracted, cDNA synthesized and the relative abundance of CRG1 transcript was analyzed by qRT-PCR. Data are the mean of at least three independent experimental replicates, and error bars represent the standard deviation. (C) Point mutations in conserved residues of the Crg1 methyltransferase domain reduce cantharidin tolerance. Site-specific mutations in the conserved motifs of methyltransferase domain are shown with the arrows. Mutated CRG1 ORFs were cloned under GAL1 promoter and transformed into crg1Δ/Δ cells. The transformants were grown in SD-URA with raffinose (2%) to mid-exponential phase and induced with galactose (2%). The fitness of point mutants based on saturation at final OD was assessed in the presence of cantharidin (6 µM). (D) A plot comparing the transcriptome profiles of crg1Δ/Δ and CRG1-overexpressing crg1Δ/Δ mutants. Exponentially grown cells were treated with cantharidin (250 µM) for 1 hour or DMSO, total RNA was extracted and synthesized cDNA was hybridized to Affymetrix Tiling arrays. Significantly different GO Biological processes are listed in Table S1. (E) Diagram showing that the methionine biosynthesis is tightly linked to SAM cycling in methylation reactions coordinated by methyltransferases. The genes that are transcriptionally different between cantharidin-resistant and sensitive mutant are shown in red. STR3 (P-value <0.0057), MET17 (P-value <0.0036), MET6 (P-value <0.012), SAM1 (P-value <0.02), SAH1 (P-value <0.03), MMP1 (P-value <0.04), SAM2 (P-value <0.056) (Table S2).