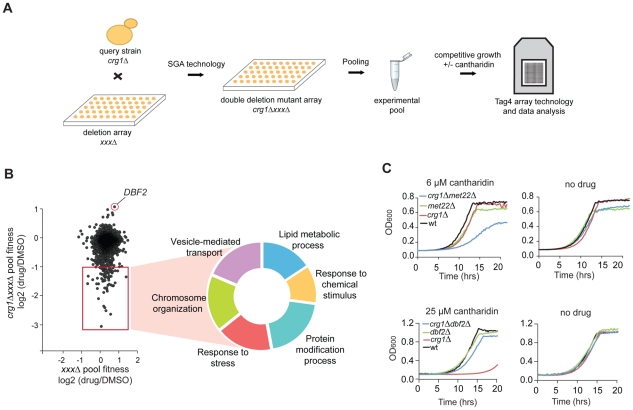
Figure 4. Characterization of cantharidin-specific genetic interactome of CRG1.
(A) Experimental scheme for analysis of cantharidin-specific genetic interactors of CRG1. Double deletion mutants crg1ΔxxxΔ generated through Synthetic Genetic Array (SGA) were pooled together and treated with cantharidin (30 µM) for 20 generations in YPD. Genomic DNA was isolated, unique strain-representative barcodes were PCR amplified, and the PCR products were hybridized to TAG4 arrays for the quantitative analysis of fitness of the mutants (see Materials and Methods for details). (B) Scatter plot representing cantharidin-gene interactions obtained from the comparative analysis of ura3ΔxxxΔ (control single deletion pool) and crg1ΔxxxΔ pools. CRG1-dependent interactors are highlighted in the red square. The hits are obtained from the averaged datasets (n = 6 for crg1ΔxxxΔ pools and n = 4 for ura3ΔxxxΔ). Significant negative genetic interactors were categorized according to their biological processes (P-value <0.002 before multiple testing correction) (Table S3). (C) Representative growth curves for the top hits (sensitive and resistant) that genetically interact with CRG1 in the presence of cantharidin. Cells were grown in YPD media with and without cantharidin. met22 and dbf2 deletion strains were treated with cantharidin to test their sensitivity and resistance, respectively.