Abstract
Motivation: The area under the receiver operating characteristic (ROC) curve (AUC), long regarded as a ‘golden’ measure for the predictiveness of a continuous score, has propelled the need to develop AUC-based predictors. However, the AUC-based ensemble methods are rather scant, largely due to the fact that the associated objective function is neither continuous nor concave. Indeed, there is no reliable numerical algorithm identifying optimal combination of a set of biomarkers to maximize the AUC, especially when the number of biomarkers is large.
Results: We have proposed a novel AUC-based statistical ensemble methods for combining multiple biomarkers to differentiate a binary response of interest. Specifically, we propose to replace the non-continuous and non-convex AUC objective function by a convex surrogate loss function, whose minimizer can be efficiently identified. With the established framework, the lasso and other regularization techniques enable feature selections. Extensive simulations have demonstrated the superiority of the new methods to the existing methods. The proposal has been applied to a gene expression dataset to construct gene expression scores to differentiate elderly women with low bone mineral density (BMD) and those with normal BMD. The AUCs of the resulting scores in the independent test dataset has been satisfactory.
Conclusion: Aiming for directly maximizing AUC, the proposed AUC-based ensemble method provides an efficient means of generating a stable combination of multiple biomarkers, which is especially useful under the high-dimensional settings.
Contact: lutian@stanford.edu
Supplementary Information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Given that there are multiple biomarkers and a binary response of interest (e.g. case and control), it is often of substantial interest to combine the biomarkers to form a ‘strong’ scoring system for the differentiation of cases from controls. While the choice of the predictive measure is not unique, the most appealing choice is the area under the receiver operating characteristic (ROC) curve (AUC) in the case–control study (Pepe, 2003; Zhou et al., 2002). For finite samples, AUC is simply the non-parametric two-sample Mann–Whitney U test statistics. Unlike the measures such as misclassification rate, the AUC reflects the intrinsic predictive value of a score in that it does not depend on the prevalence of the cases and thus is invariant under the case–control sampling. Therefore, it is natural to combine biomarkers by maximizing the AUC under ROC curve (Ma and Huang, 2005, 2007; Pepe et al., 2006; Ye et al., 2007; Zhou et al., 2011). However, it is notoriously difficult to maximize the AUC numerically since the objective function is neither continuous nor convex. Ad hoc methods have been proposed to tackle the numerical problem. For example, sigmoid function has been used to approximate the indicator function used in calculating AUC (Ma and Huang, 2007). However, the smoothed objective function may still have multiple local maximums, with no guarantee of locating the global maximizer by using the commonly used numerical algorithms. In view of these challenges, we propose a class of ensemble methods aiming for maximizing AUC with multiple biomarkers. Specifically, we introduce a class of convex surrogate loss functions to approximate the non-convex AUC, greatly facilitating computation and optimization.
2 METHODS
2.1 Surrogate loss functions
Assume that X1,…, Xn are n independently identically distributed (i.i.d) copies of p-dimensional random vector X, representing, for example, p biomarkers for cases, and Y1,…, Ym are m i.i.d copies of p-dimensional random vector Y for controls. Suppose that we want to construct a score as a linear combination of the p biomarkers with the aim of maximizing the AUC under ROC curves. Specifically, we want to find a vector β to maximize the objective function
Ideally, we would want the score for cases to be higher than that for controls, which yields 1 for the AUC and completely differentiates cases and controls. However, several challenges are prominent. First, since the objective is invariant in β→c0β for arbitrary c0>0, there is no unique maximizer for the objective function. One often needs to subjectively select an anchor biomarker with its weight in the linear combination being one and maximize the remaining p−1 components in β. The performance of the score heavily depends on the selection of the anchor biomarker. Second, even with a given anchor biomarker the objective function is still neither continuous nor concave and therefore it is very likely that conventionally used optimization iterations have been trapped around local maximum points depending on the subjectively selected initial point.
We are now in a position to propose a method addressing these two challenges. By noting that 1−AUC(β) can be interpreted as the misclassification rate of using the binary rule β′(Xi−Yj)>0 to classify a binary response always taking value 1, we may borrow the popular classification approaches aimed for minimizing the misclassification rate in the data mining literature (Friendman et al., 2000; Hastie and Zhu, 2006). Specifically, instead of directly maximizing AUC(β) or equivalently minimizing 1−AUC(β), it is sensible to minimize a surrogate loss function. We propose the following two surrogate functions
and
which correspond to the negative log likelihood function raised in the conditional logistic regression and hinge loss function used in support vector machine, respectively, where x+=xI(x>0). Since these two functions are continuous convex function, their minimizers are well defined and can be reliably located. Numerically, it amounts to replace the indicator function I(x<0) by log{1+exp(−x)} and (1−x)+, respectively. Previous data mining literature reveals that algorithms minimizing such surrogate loss functions often result in models with good performance in minimizing the misclassification rate. Analogously, the estimated scores tend to render satisfactory AUCs.
With moderate dimension p, while M1(β) can be minimized via the scoring algorithm in fitting a generalized linear model, we can use linear programming technique to minimize M2(β). Specifically, it is equivalent to
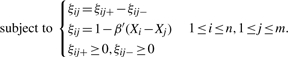 |
Several advantages of the proposal are obvious. First, minimizing either M1(β) or M2(β) does not require selecting an anchor biomarker a priori, which is especially appealing for high-dimensional case. Second, in the limiting case, the maximizer of the AUC under the ROC curve
is g(·)=m{f1(·)/f0(·)} for any strictly monotone increasing function m(·), where f1(·) and f0(·) are the underlying density functions of X and Y, respectively (Jin and Lu, 2008). The minimizer of
is log{f1(·)/f0(·)} and thus minimizing E{M1(g)} is equivalent to maximizing the AUC under the ROC curve. The minimizer of
is more complicated. Numerical studies point that the minimizer may also be a monotone transformation of f1(·)/f0(·), as opposed to the conventional support vector machine whose solution approximates the optimal decision boundary.
2.1.1 Adaptive generalizations
As neither M1(β) nor M2(β) provides a good approximation to 1−AUC(β) (indeed no convex functions accurately approximate the indicator function), we employ an iterative algorithm to approximately minimize 1−AUC(β). More specifically, given that
we expect that the minimizer of ∑ni=1 ∑mj=1 mc[β′(Xi−Yj) approximates that of 1 − AUC(β) for small c > 0. Figure 1 illustrates the 0−1 loss function against log{1+exp(−x)}, the hinge loss function and mc(·) with different cs. Noting that mc(β′x) = m1{(β/c)′x}, the scoring system minimizing mc(β′x) is equivalent to that minimizing m1(β′x). Thus, we may employ the following adaptive algorithm
- Set the initial
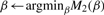
- Update β as
where the minimization can be solved via linear programming technique.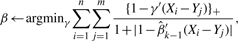
Repeat Step (2) until convergence or the number of iteration reaches a pre-specified number.
Fig. 1.
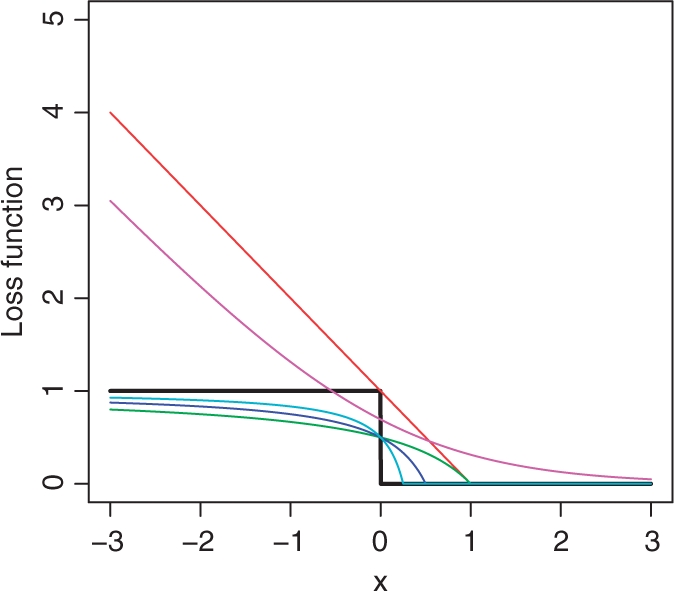
mc(x) against the 0–1 loss function. The 0−1 loss function I(x<0) (black); hinging loss function (red); green: m1(x); m0.5(x) (blue); m0.25(x) (cyan); log{1+exp(−x)} (purple).
Our numerical results (reported in the Supplementary Material) show that the adaptive iteration may increase the resulting AUC especially when there are potential outliers. The robustness of the method is not a surprise, because the influence from individual observations on the objective function via mc(x) is always bounded. Furthermore, we find that one or two iterations often suffice to harvest most of the gain in maximizing AUC(β) and thus, in general, there is no need to continue the iteration until convergence.
2.1.2 Extension to survival outcomes
When the outcome is survival time subject to potential right censoring, the c− index as the generalized AUC is often computed as
where  are n i.i.d copies of
are n i.i.d copies of 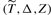 ,
, 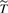 is the minimum of the censoring and failure times, Δ is the binary censoring indicator and Z is the covariate vector (Cai and Cheng, 2008). Similarly, one may find β by minimizing
is the minimum of the censoring and failure times, Δ is the binary censoring indicator and Z is the covariate vector (Cai and Cheng, 2008). Similarly, one may find β by minimizing
and
the counterparts of M1(β) and M2(β), respectively. Indeed, the log-partial likelihood function also can be viewed as surrogate to the c−index in that both
may serve as a surrogate to
This may explain that the Cox model-based c−index is often high.
2.2 Regularization for high-dimensional covariates
When p is high, the proposed surrogate loss function can be conveniently regularized for feature selection. While many regularization methods can be used, we hereafter pursue the popular lasso regularization for illustration (Tibshirani, 1996). Specifically, we propose to minimize
where β=(β1,…, βp)′ and |β|=∑j=1p |βj|. For M1(β), one may use the following iterative coordinate descending algorithm to minimize the objective function with a given λ
Set an initial estimator β.
- Update β
where Wij=pij(1−pij),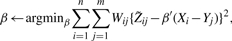
and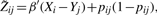
for i=1,…, n and j=1,…, m. The standard coordinate descending algorithm can be used to minimize the weighted L2 loss function in this step (Friedman et al., 2010).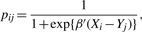
Repeat Step 2 until convergence.
The coordinate descending algorithm is not directly applicable for the non-differentiable surrogate loss function M2(β). However, since the objective function M2(β) is convex and piecewise linear, the exact solution path with λ varying from ∞ to 0 is also piecewise linear and can be computed using the generalized LARS algorithm (Cai et al., 2009; Rosset and Zhu, 2007). When the dimension p or sample size n is high, the computation is demanding due to dense joints in the solution path. As a remedy, we propose a forward stagewise algorithm that generates an approximate solution path of β (Friedman and Popescu, 2004).
Set an initial estimator β=0 and small positive number ϵ>0.
- At step k=1,…, identify the coordinate j with the largest decrease
and update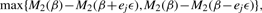
where ϵ>0 is an small constant selected a priori, ej is a p-dimensional vector with all the components being zero except the j-th component, which is 1 and sj = 2I{M2(β+ejε)<M2(β−ejε)}−1.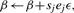
Repeat Step 2 until the number of non-zero components of β reaches a prespecified number or the AUC(β) becomes 1.
When the exact lasso solution is desirable, one may employ an ad hoc two-stage approach. Specifically, one may first implement the aforementioned forward stagewise algorithm to screen informative features. The forward stagewise algorithm stops when the number of selected features reaches a prespecified maximum number of biomarkers to be used for constructing the score in practice, say 30. At the second step, the exact lasso solution path can be computed with only the selected features. In either case, the penalty parameter can be selected via cross-validation. The objective function used in the cross-validation can be either Mj(β) itself or the AUC under ROC curve.
3 SIMULATION
Extensive simulations are conducted to examine the finite sample performance of the proposed method. We generate the covariates {X1,…, Xn} and {Y1,…, Ym} from the following models:
(multivariate normal)
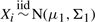 and
and 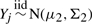 , where Xi and Yj are p-dimensional random vectors.
, where Xi and Yj are p-dimensional random vectors.(normal mixture)
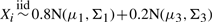 and
and 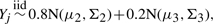 i.e. 20% of the markers values in both cases and controls are contaminated by a common error distribution.
i.e. 20% of the markers values in both cases and controls are contaminated by a common error distribution.(multivariate log-normal)
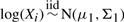 and
and 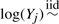
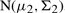 .
.(log-normal mixture)
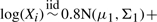
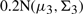 and
and 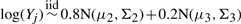 .
.
In the above settings, we let μ1=(1, 0, 0,…, 0)′, μ2=(0, 1, 0,…, 0)′, μ3=(1, 1, 1, 0,…, 0)′,
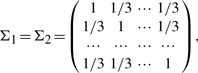 |
Σ3=5Ip, Ip is the identity matrix. We considered several configurations of n, m and p to investigate the operational characteristics of the proposed method.
First, we examine the scenario where the number of covariates is low relative to the sample size. To this end, we let p=3 and n=m=50. For each generated dataset, we construct a linear combination of the covariates as a score differentiating cases from controls, where the weights of the linear combinations are estimated by minimizing (i) M1(β) (ii) M2(β) (iii) the loss function
| (1) |
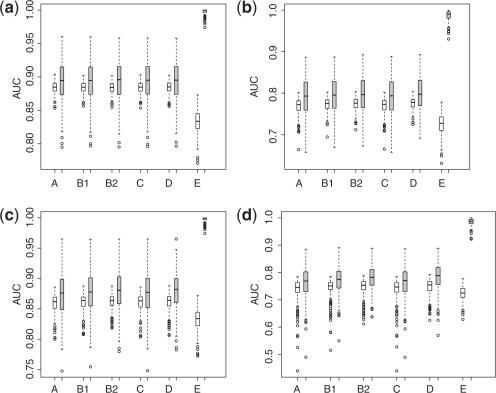
proposed in Ma and Huang (2007) and (iv) by fitting a regular logistic regression. We also implement the popular ada-boosting with 300 iterations using the simple stump as the base classifier (Friendman et al., 2000). The continuous class probability based on ada-boosting trained ensemble is used to generate the ROC curve. In minimizing S(β), we first identify the ‘anchor covariate’ with the most significant p value from t-test comparing the covariate distribution between cases and controls and set its regression coefficient at +1 or −1 depending on the sign of the t-statistics. The σ in S(β) is then selected as 20% of the mean group difference of the anchor covariate as suggested in Ma and Huang (2007). We then calculate the AUCs in an independent test set consisting of 2000 cases and 2000 controls for all the five constructed scores. The boxplots of AUCs over 250 replications in each setting are plotted in Figure 2. The AUCs in the training sets are higher than their counterparts in the validation sets as expected. In most cases including the first setting, where the logistic regression estimates the optimal combination in terms of maximizing the AUC, the scores based on M1(β), M2(β), S(β) and logistic regression perform similarly in terms of AUC in the validation sets. In general, the score based on ada-boosting has the lowest AUC, which could be due to overfitting indicated by the high AUCs in the training set. Furthermore, the score based on the one-step adaptive hinge loss function performs similar or slightly superior to that based on hinge loss function itself.
Fig. 2.
Empirical AUC in both training and validation sets for different methods [A: M1(β); B1: M2(β); B2: one-step adaptive M2(β); C: S(β); D: lasso-regularized logistic regression; E: ada-boosting using stumps with 300 iterations] with low-dimensional covariates. Gray box: training set; empty box: validation set. (a) Simulation setting I (normal); (b) simulation setting II (normal mixture); (c) simulation setting III (log-normal); (d) simulation setting IV (log-normal mixture).
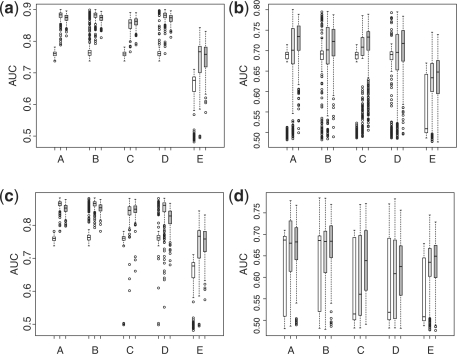
Second, we have examined the performance of the proposed method for covariates with moderate dimension. In this case, we let p=200 and n=m=50 and the lasso regularization is used for selecting the important features in logistic regression. The forward stagewise algorithm similar to that presented in Section 2.2 for minimizing M2(β) is also used to minimize S(β). We choose the popular lasso penalty mainly for the purpose of fair comparison, i.e. evaluating the relative performance of various methods under similar regularization schemes. The boxplots of AUCs in independent test sets over 250 replications are plotted in Figure 3. In general, the scores based on Mj(β) perform better than that based on the alternatives in terms of average AUC in the test sets. Furthermore, the AUCs from scores constructed via Mj(β) also tend to have smaller variability than their counterparts. In the most challenging fourth setting, the empirical average AUC in the test sets is 0.66 for score minimizing M1(β), 0.66 for score minimizing the hinge loss, M2(β), 0.60 for scores minimizing S(β), 0.60 for score from the logistic regression fitting and 0.63 for score from ada-boosting using three markers. An increase from 0.60–63 to 0.66 in the AUC is often considered non-trivial in clinical practice.
Fig. 3.
Empirical AUC in the validation sets for different methods [A: M1(β); B: M2(β); C: S(β); D: lasso-regularized logistic regression; E: ada-boosting using stumps with 300 iterations] with moderate dimensional covariates. Empty box: AUC with one selected covariate; gray box: AUC with three selected covariates; dark gray box, AUC with 10 selected covariates. (a) Simulation setting I (normal); (b) simulation setting II (normal mixture); (c) simulation setting III (log-normal); (d) simulation setting IV (log-normal mixture).
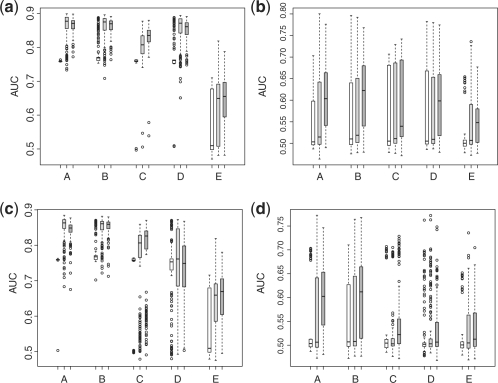
Lastly, we have examined the cases for high-dimensional covariate. Here, we let p=20 000 and n=m=50. To save computational time, the ada-boosting is only applied to top 500 features selected based on significance levels of t-test comparing cases and controls in the training set. The simulation results are presented in Figure 4. For the high-dimensional covariates, the relative performance of the proposed methods is even better than that in the previous case where p=200. For example, in the third setting, the empirical average AUC in the test sets is 0.85 for score minimizing M1(β), 0.85 for score minimizing the hinge loss, 0.76 for scores minimizing S(β), 0.74 for score from the logistic regression fitting and 0.64 for score from ada-boosting using three markers. Similarly, in the fourth setting, the empirical average AUC in the test sets are 0.56, 0.56, 0.53, 0.53 and 0.53 for aforementioned five methods.
Fig. 4.
Empirical AUC in the validation sets for different methods [A: M1(β); B: M2(β); C: S(β); D: lasso-regularized logistic regression; E: ada-boosting using stumps with 300 iterations] with high-dimensional covariates. Empty box: AUC with one selected covariate; gray box: AUC with three selected covariates; dark gray box, AUC with 10 selected covariates. (a) Simulation setting I (normal); (b) simulation setting II (normal mixture); (c) simulation setting III (log-normal); (d) simulation setting IV (log-normal mixture).
4 ANALYSIS OF THE BONE MINERAL DENSITY STUDY
We apply the proposed method to a dataset (Reppe et al., 2010) arising from a study that recruited 301 non-related post-menopausal ethnic Norwegian women at the Lovisenberg Deacon Hospital. Among them, bone mineral density (BMD) and gene expression levels (Affymetric array) were measured for 84 women. Since low BMD is associated with higher fracture rates (Cooper, 1997), it is of interest to identify a linear combination of gene expression levels to differentiate the osteopenia or osteoporosis (low BMD) from normal among post-menopausal women. Bone biopsies show that there are 39 from 84 women having osteopenia or osteoporosis. All the normalized gene expression level are log-transformed. After screening out ~25% probesets with lowest variation, we have 40 411 probesets for each patient. We randomly split the data into training and validation sets and apply the proposed method to the training set consisting of 26 cases and 30 controls. For the purpose of comparisons, we also construct gene scores based (1) S(β) proposed in Ma and Huang (2007) (2) lasso-regularized logistic regression and (3) ada-boosting using stumps as base classifiers. The anchor gene and σ in S(β) are determined using the same method as that presented in the simulation study. To save computational time, ada-boosting is only applied to the top 2000 genes according to their significance level in t-test comparing average gene expression levels between cases and controls. With the estimated scores based on M1(β), M2(β), S(β), the regularized logistic regression and ada-boosting, we examine their corresponding AUC in the validation set. The results are shown in Figure 5. It can be seen that in general scores based on M1(β) and M2(β) yield higher AUC than that based on S(β), the commonly used logistic regression and ada-boosting with the same number of covariates in the validation set. In Figure 5, we also plot the AUC in the training set. Since the number of covariates is much higher than the sample size, the maximum AUC (AUC = 1) corresponding to complete separation between case and control in the training set is reached with 20–30 covariates for all the methods. The highest AUCs for scores based on S1(β), regularized logistic regression and ada-boosting are 0.764, 0.687 and 0.728, respectively, while the highest AUC is 0.708 for score based on M1(β) and 0.764 for score based on M2(β). As a reference, the AUC for age is only 0.669 in this cohort. Furthermore, while the optimal scores with M1(β) and M2(β) use 9 and 13 genes, respectively, their counterparts based S(β), regularized logistic regression and ada-boosting use 37, 12 and 45 genes, respectively. These comparisons suggest that the genes score based on M2(β) possesses the best combination of sparsity and prediction performance: it attains the highest AUC in the validation set with only 13 genes. The gene lists selected by these methods are heavily overlapping. For example, there are seven common genes shared by at least three out of four linear combinations constructed based M1(β), M2(β), S(β) and logistic regression. The probe set AFFX-M27830_M_at, which is shared by all four scores, is a member of the eight core genes reported by (Reppe et al., 2010). Furthermore, gene SOST (Affymetrix ID 223869_at) shared by scores based on M1(β), M2(β) and logistic regression is also a member of the eight core genes explaining the variation of BMD and sits in the ‘center’ of the constructed intermolecular network sharing significant associations reported in the original paper (Reppe et al., 2010). The selected genes and their corresponding weights are summarized in Table 1, where the weights are standardized such that the probe set AFFX-M27830_M_at has the unit weight for comparison purpose. One interesting and reassuring observation is that signs of all non-zero weights were consistent across methods.
Fig. 5.
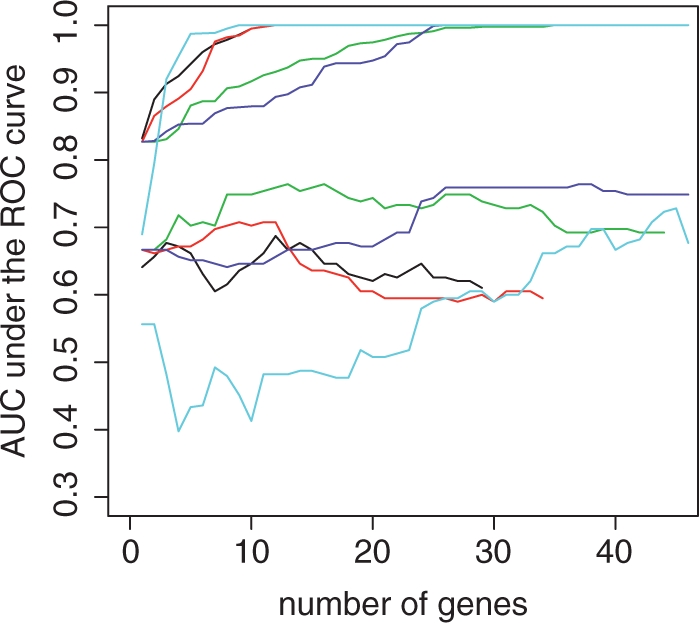
AUC of scores for differentiating the women having low and normal BMD. Thick lines: AUC in validation set; thin lines: AUC in training set; red: score based on M1(β); green: score based on M2(β); blue: score based on S(β); black: score based on logistic regression; cyan: score based on ada-boosting.
Table 1.
The estimated scores for differentiating low and normal BMD
| Affymatrix ID | Weights |
|||
|---|---|---|---|---|
| M1(β) | M2(β) | S(β) | LR | |
| AFFX-M27830_M_at | 1 | 1 | 1 | 1 |
| 211769_x_at | 0 | 0 | 0 | 0.946 |
| 215887_at | 0 | 0 | 0 | 1.915 |
| 217761_at | 0 | 0 | 0 | −0.447 |
| 220900_at | 0 | 0 | 0 | 1.073 |
| 223869_at | 0.587 | 0.159 | 0 | 0.112 |
| 227405_s_at | 0.055 | 0 | 0 | 0.024 |
| 231599_x_at | 0 | 0 | 0 | −0.175 |
| 235102_x_at | 0.983 | 0.113 | 0 | 2.822 |
| 237739_at | 0.779 | 0.093 | 0 | 0.896 |
| 238020_at | 0 | 0 | 0 | −1.625 |
| 239498_at | 1.161 | 0.185 | 0 | 1.146 |
| 206742_at | −0.068 | −0.06 | −0.026 | 0 |
| 222735_at | −0.639 | −0.185 | −0.082 | 0 |
| 244035_at | 0.692 | 0.013 | 0 | 0 |
| 219747_at | 0 | 0.007 | 0 | 0 |
| 235439_at | 0 | 0.033 | 0 | 0 |
| 238705_at | 0 | 0.212 | 0 | 0 |
| 238946_at | 0 | 0.06 | 0.02 | 0 |
| 1552477_a_at | 0 | −0.02 | 0 | 0 |
| 206273_at | 0 | 0 | 0.008 | 0 |
| 206307_s_at | 0 | 0 | 0.006 | 0 |
| 206326_at | 0 | 0 | 0.038 | 0 |
| 207369_at | 0 | 0 | −0.036 | 0 |
| 210045_at | 0 | 0 | −0.002 | 0 |
| 210174_at | 0 | 0 | −0.036 | 0 |
| 214412_at | 0 | 0 | −0.004 | 0 |
| 215196_at | 0 | 0 | −0.008 | 0 |
| 215431_at | 0 | 0 | 0.002 | 0 |
| 219566_at | 0 | 0 | −0.01 | 0 |
| 220554_at | 0 | 0 | 0.09 | 0 |
| 220584_at | 0 | 0 | −0.012 | 0 |
| 221631_at | 0 | 0 | −0.01 | 0 |
| 227440_at | 0 | 0 | −0.078 | 0 |
| 229201_at | 0 | 0 | −0.108 | 0 |
| 230349_at | 0 | 0 | −0.01 | 0 |
| 230839_at | 0 | 0 | −0.024 | 0 |
| 231231_at | 0 | 0 | 0.01 | 0 |
| 231468_at | 0 | 0 | −0.112 | 0 |
| 231759_at | 0 | 0 | −0.106 | 0 |
| 231828_at | 0 | 0 | −0.004 | 0 |
| 232114_at | 0 | 0 | 0.09 | 0 |
| 234259_at | 0 | 0 | 0.012 | 0 |
| 234421_s_at | 0 | 0 | −0.036 | 0 |
| 234604_at | 0 | 0 | 0.058 | 0 |
| 241736_at | 0 | 0 | 0.048 | 0 |
| 243673_at | 0 | 0 | −0.016 | 0 |
| 243889_at | 0 | 0 | −0.068 | 0 |
| 244338_at | 0 | 0 | 0.022 | 0 |
| 1553027_a_at | 0 | 0 | −0.008 | 0 |
| 1556803_at | 0 | 0 | 0.044 | 0 |
| 1556938_a_at | 0 | 0 | 0.048 | 0 |
| 1560779_a_at | 0 | 0 | 0.032 | 0 |
LR, logistic regression.
We also repeat analysis based on other random training test splitting and obtain similar results.
5 DISCUSSION
Motivated by recent advances in data mining, we have proposed a class of methods combining biomarkers to construct a scoring system, boosting the resulting AUC under the ROC curve, a prevalence-free summarization of intrinsic predictive values of a continuous score. The method is easily adapted to high-dimensional cases, wherein one may need to identify informative features from thousands of candidate biomarkers. In high-dimensional case, we propose to apply lasso regularization to yield a parsimonious combination maximizing the AUC. On the other hand, lasso is neither the unique nor the universally optimal regularization method for analyzing high-dimensional data. Due to the convexity of the proposed loss function, it is straightforward to couple Mj(β) with other penalty functions such as elastic net, adaptive lasso and SCAD, which may have superior performance to simple lasso in specific settings (Zou, 2006; Zou and Hastie, 2005; Zou and Li, 2008). The key proposal is to target a convex surrogate loss function instead of a discontinuous Mann–Whitney rank statistic.
While in this article, we have focused on the hinge loss function (corresponding to the 1 − norm support vector machine), our results can be extended to accommodate other versions of SVM loss functions, such as
for any given α≥1. Another alternative is the exponential function used in boosting algorithm
Finally, while the AUC under the entire ROC curve is a useful global measure, the AUC under partial ROC curve has recently emerged as a useful problem-specific measure in practice (Komori and Equchi, 2010). Therefore, an numerically efficient algorithm combining multiple biomarkers to maximize the AUC under partial ROC curves or sensitivity for given specificity level is worth further investigations.
Funding: R01 HL089778-04 (to L. Tian) and R01 CA95747 (to Y. Li) from National Institute of Health.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Cai T, Cheng S. Robust combination of multiple diagnostic tests for classifying censored event times. Biostatistics. 2008;9:216–233. doi: 10.1093/biostatistics/kxm037. [DOI] [PubMed] [Google Scholar]
- Cai T, et al. Regularized estimation for the accelerated failure time model. Biometrics. 2009;65:394–404. doi: 10.1111/j.1541-0420.2008.01074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper C. The crippling consequences of fractures and their impact on quality of life. Am. J. Med. 1997;103:12S–17S. doi: 10.1016/s0002-9343(97)90022-x. [DOI] [PubMed] [Google Scholar]
- Friedman J, Popescu B. Gradient directed regularization for linear regression and classification. Technical Report. Department of Statistics, Stanford University; 2004. [Google Scholar]
- Friedman J, et al. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softwr. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- Friendman J, et al. Additive logistic regression: a statistical view of boosting. Ann. Stat. 2000;28:337–407. [Google Scholar]
- Hastie T, Zhu J. Discussion of “support vector machines with applications” by Javier Moguerza and Alberto Munoz. Stat. Sci. 2006;21:352–357. [Google Scholar]
- Jin H, Lu Y. A procedure for determining whether a simple combination of diagnostic tests may be noninferior to the theoretical optimum combination. Med. Decis Making. 2008;28:909–916. doi: 10.1177/0272989X08318462. [DOI] [PubMed] [Google Scholar]
- Komori O, Equchi S. A boosting method for maximizing the partial area under the ROC curve. BMC Bioinformatics. 2010;11:314–330. doi: 10.1186/1471-2105-11-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Huang J. Regularized roc method for disease classification and biomarker selection with microarray data. Bioinformatics. 2005;21:4356–4362. doi: 10.1093/bioinformatics/bti724. [DOI] [PubMed] [Google Scholar]
- Ma S, Huang J. Combining multiple markers for classification using ROC. Biometrics. 2007;63:751–757. doi: 10.1111/j.1541-0420.2006.00731.x. [DOI] [PubMed] [Google Scholar]
- Pepe M. The Statistical Evaluation of Medical Tests for Classification and Prediction. Oxford: Oxford University Press; 2003. [Google Scholar]
- Pepe M, et al. Combining predictors for classification using the area under the receiver operating characteristic curve. Biometrics. 2006;62:221–229. doi: 10.1111/j.1541-0420.2005.00420.x. [DOI] [PubMed] [Google Scholar]
- Reppe S, et al. Eight genes are highly associated with bmd variation in postmenopausal caucasian women. Bone. 2010;46:604–612. doi: 10.1016/j.bone.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Rosset S, Zhu J. Piecewise linear regularized solution paths. Ann. Stat. 2007;35:1012–1030. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc., Ser. B. 1996;58:267–288. [Google Scholar]
- Ye J, et al. On the analysis of glycomics mass spectrometry data via the regularized area under the ROC curve. Bioinformatics. 2007;8:477–488. doi: 10.1186/1471-2105-8-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, et al. Statistical Methods in Diagnostic Medicine. New York: John Wiley & Sons, Inc.; 2002. [Google Scholar]
- Zhou X, et al. Variable selection using the optimal roc curve: An application to a traditional chinese medicine study on osteoporosis disease. Stat. Med. 2011 doi: 10.1002/sim.3980. [Epub ahead of print, doi:10.1002/sim.3980] [DOI] [PubMed] [Google Scholar]
- Zou H. The adaptive lasso and its oracle properties. J. Am. Stat. Assoc. 2006;101:1418–1429. [Google Scholar]
- Zou H, Hastie T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B. 2005;67:301–320. [Google Scholar]
- Zou H, Li R. One-step sparse estimates in nonconcave penalized likelihood models. Ann. Stat. 2008;36:1509–1533. doi: 10.1214/009053607000000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.