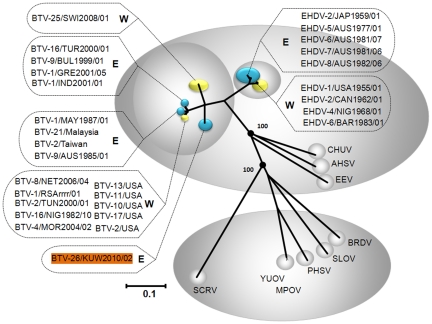
Figure 1. Neighbour-joining tree showing relationships between VP3[T2] of KUW2010/02 with other orbiviruses.
KUW2010/02 showed up to 76.6%/88.9% nt/aa identity in Seg-3/VP3[T2] with other BTV strains confirming that it is an isolate of BTV. Accession numbers and further detail of the sequence and viruses used are included in Table 1. The tree was constructed using distance matrices, generated using the p-distance determination algorithm in MEGA 5 (500 bootstrap replicates) [48]. The trees shown in Figures 2 and 3 were drawn using same parameters. The scale bar indicates the number of substitutions per site. Values at the nodes indicate bootstrap confidence. Epizootic haemorrhagic disease virus (EHDV), Bluetongue virus (BTV), Equine encephalosis virus (EEV), African horse sickness virus (AHSV), Chuzan virus (CHUV), St. Croix River virus (SCRV), Yunnan orbivirus (YUOV), Middle point orbivirus (MPOV), Peruvian horsesickness virus (PHSV), Broadhaven virus (BRDV), Stretch Lagoon Orbivirus (SLOV). Eastern and western isolates of EHDV and BTV are shown in blue and yellow respectively. Seg-3 accession numbers used for comparative analyses: AM745079, AM745029, AM745039, AM745049, AM745059, AM744979, AM744999, AM745019, AM745069, NC_005989, AF021236, FJ183386, M87875, NC_012755, NC_007749, NC_007657, EF591620, NC_005998, DQ186827, DQ186797, DQ186822, DQ186811, DQ186816, AF529047, AY493688, DQ186790, AM498052, DQ186792, DQ186826, DQ186819, DQ186817, L19969, L19968, NC 006014, AF017281, L19967.