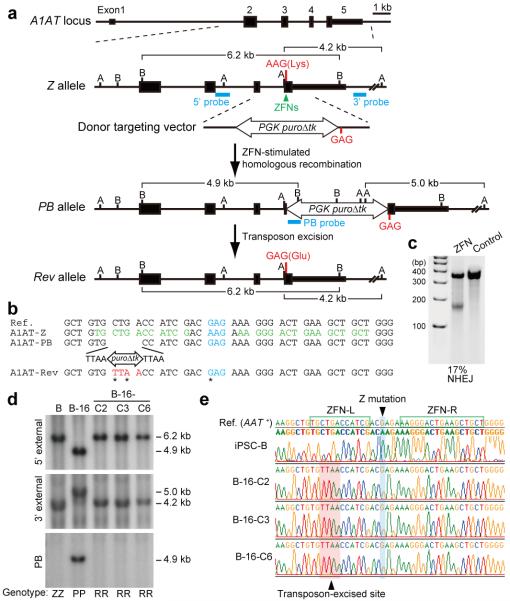
Figure 2. Correction of the Z mutation in A1ATD-hIPSCs.
a, The strategy for precise genome modification using ZFNs and the piggyBac transposon. Top line, structure of the A1AT gene; blue lines, Southern blot probes; thin and thick boxes, non-coding and coding exons, respectively; open arrow, piggyBac transposon; B, BamHI; A, AflIII. b, Sequences of wild-type (Reference), Z, PB, and Rev alleles. Amino acid position 342 (blue), recognition sites for ZFNs (green), piggyBac excision site (red) are shown. Sequence changes in Rev allele from Z allele were indicated by asterisks. c, Surveyor nuclease assay showing the cleavage of Z mutation in ZFNs-transfected K562 cells. Non-transfected cells were used as a control. d, Southern blot analysis showing bi-allelic piggyBac insertion (B-16) and bi-allelic excision (B-16-C2, -C3 and -C6) during correction of the A1ATD-hIPSCs line B. Genomic DNA was digested by BamHI (5′ and PB probes) or AlfIII (3′ probe). Genotype: ZZ, homozygous for Z allele; PP, homozygous for insertion of piggyBac; RR, homozygous for reverted allele. e, Sequence analysis showing correction of Z mutation in 3 corrected hIPSC lines. Wild-type sequence (top line) and A1ATD-hIPSC (second line).