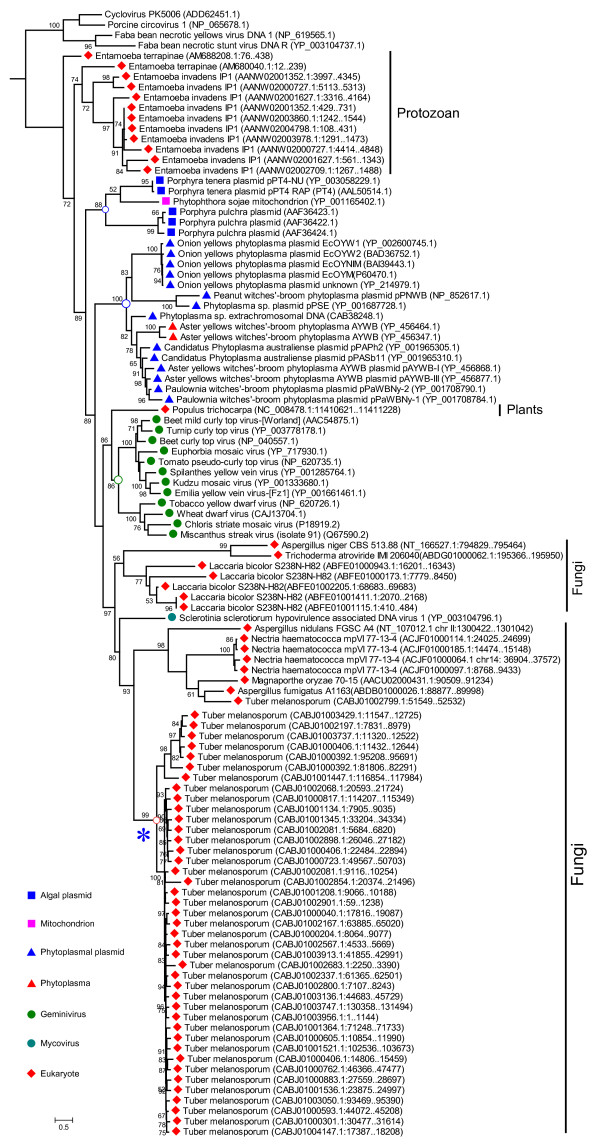
Figure 4.
Phylogeny of geminiviral Rep-like sequences from eukaryotes, known viruses, plasmids and phytoplasma. The phylogenetic tree was built using PhyML-mixtures based on a multiple sequence alignment generated using COBALT with the Constraint E-value parameter setting to 0.1. This tree was rooted with circoviruses and nanoviruses. The topology of blue asterisk marked clade was evaluated independently. Only p-values of the approximate likelihood ratios (SH-test) > 0.5 (50%) are indicated. scale bars correspond to 0.5 amino acid substitutions per site. Sequence accession numbers are given for each sequence.