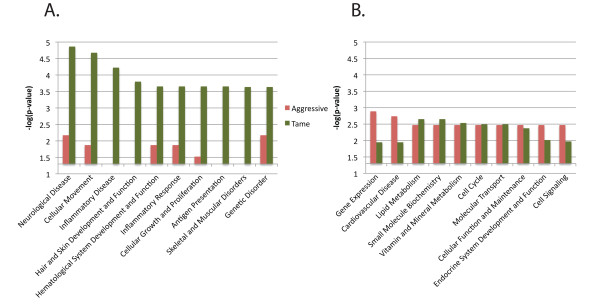
Figure 5.
Biofunction groups of genes expressed differentially between tame and aggressive foxes. Expression differences between the tame and aggressive individuals were evaluated as the proportionate read count per gene. Genes with at least a 2-fold difference in expression (at p < 0.05) were sorted into Biofunction groups by Ingenuity IPA (Version 8.6). Biofunction groups were then ranked by the negative logarithm of the biofunction p-value, which estimates the probability that the biofunction group is over-represented in the set of differentially expressed genes. The vertical axis gives the statistical support for each bioinformatic group being upregulated in the tame (green) or aggressive individual (red) (a). The 10 highest ranked biofunction groups representing genes with higher expression in the tame individual. (b). The 10 highest ranked biofunction groups representing genes with higher expression in the aggressive individual.