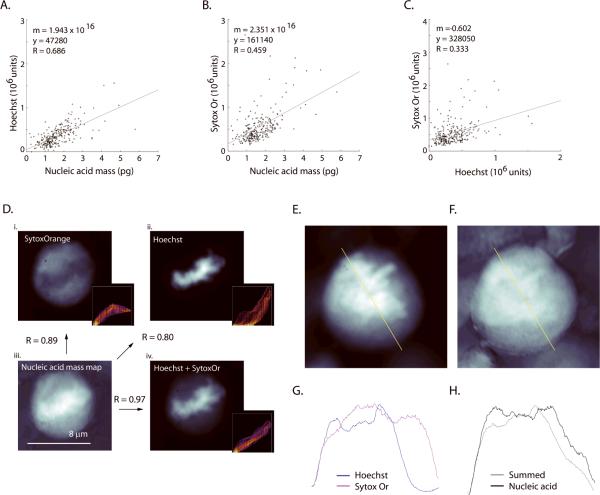
Figure 6. Microstructural comparisons reveal differences between mass map and nuclear stains.
(A) Moderate correlation (R=0.69, n = 260 cells) is seen to the integrated Hoechst signal, (B) weaker correlation (R=0.46) to Sytox Orange (SytoxOr) and (C) (as expected) poor correlation (R=0.33) between Hoechst and Sytox Orange channels were observed. (D) Global correlation of mass map to stains. The same cell is imaged at 100× magnification with Sytox Orange (i, 1 μM) and Hoechst 33342 (ii, 16.2 μM). Panel (iii) shows UV-derived nucleic-acid mass and panel (iv) as an equally-weighted linear sum of the normalized images from Hoechst and Sytox Orange. The summed image in (iv) shows higher global correlation to the nucleic acid mass map (also see Fig. 9). Arrows denote the comparisons and the associated Pearson's correlation score (`normxcorr2' function in MatLab) that are made between the panels within (D). Insets show a frequency distribution of pixel values for the nucleic acid and the fluorescence images. SytoxOr staining was performed on formaldehyde fixed cells without permeabilization or addition of RNase. A summed Hoechst+SytoxOr image (E) and nucleic acid mass map (F) of a mitotic cell. Line plots of the same pixels taken from the fluorescence images (G; blue: Hoechst, magenta: SytoxOr) and from the nucleic acid mass map and the equally weighted, summed fluorescence image (H; gray: summed image, black: nucleic acid). Line plots taken from the yellow line in A and B. It appears that the two nuclear stains cannot be summed in any combination so as to reproduce the contours of the nucleic acid mass map.