Abstract
The important role of activating Killer Immunoglobulin-like Receptors (aKIR) in protecting against cytomegalovirus (CMV) reactivation has been described previously in hematopoietic cell transplantation (HCT). More specifically, the presence of multiple aKIR and the presence of at least KIR2DS2 and KIR2DS4 in the donor genotype identified a group of HCT patients that were at low risk for CMV reactivation. However, CMV infection still occurs in patients with KIR protective genotype and the question was raised as to whether this was due to the lack of KIR expression. In this report, the expression of KIR2DS2 and 2DS4 gene, as measured by mRNA-based Q-PCR both in the donor cells and in the HCT recipient cells was studied relative to CMV reactivation. In the control samples from healthy HCT donors, the median range of for KIR2DS2 and KIR2DS4 expression was low with 35% considered null-expressers. Interestingly, KIR2DS2 and KIR2DS4 expression was elevated after HCT when compared to donor expression prior to transplant, and significantly elevated in the CMV viremic (V) compared to non-viremic (NV) HCT recipients. CMV seropositivity of donors was not associated with aKIR expression, and donor null-expression in those with KIR2DS2 or KIR2DS4 genotype did not predict for CMV reactivation in the recipient. After controlling for other transplant factors that included donor type (sibling or unrelated), transplant source -bone marrow (BM) or peripheral blood stem cells (PB) and acute GVHD grade, the result of the regression analysis of elevated KIR gene expression was found to be associated for both KIR2DS2 and KIR2DS4, with seven fold increase in risk for CMV reactivation. We speculate that the elevated aKIR expression in CMV viremic HCT recipients is either coincidental with factors that activate CMV or is initiated by CMV or cellular processes responsive to such CMV infection reactivation.
Introduction
Earlier reports have emphasized the importance of NK cells and their KIR receptor in controlling CMV reactivation after hematopoietic cell transplant (HCT). More specifically, donors with more than one activating KIR gene were associated with 65% reduction in CMV reactivation [1–2] and the same effect has been seen in kidney transplants [3]. In the HCT setting, it was determined that the number of activating KIR genes in the donors but not in the recipients, was associated with protection from CMV reactivation and that the protective effect was highest when the donor genotype contained >5 aKIR genes or at least a combination of KIR2DS2 and KIR2DS4 [4]
However, a KIR genotype profile does not correlate necessarily with concurrent expression. The question is whether expression of the aKIRs predicts protection from CMV, and, conversely, does the non-expression of these genes explain the failure of a KIR2DS2/KIR2DS4 genotype to protect from CMV infection? The expression of the donor aKIR genes KIR2DS2 and KIR2DS4 was evaluated in HCT recipients after allogeneic stem cell transplantation. The effect of HCT regimen as well as cell reconstitution with aKIR gene expression was analyzed to answer these questions and to test whether expression levels are modulated by post-HCT events such as CMV reactivation or GVHD. This report focuses on activating KIR2DS2 and KIR2DS4 gene expression, since this dual aKIR genotype appears to be protective, and examines quantitatively the occurrence of expression relative to CMV infection in recipients of allogeneic HCT.
While KIR expression can be detected by flow-cytometry, the available antibodies do not distinguish the activating KIR2DS2 and its linked gene, the inhibitory KIR2DL2. An mRNA-based quantitative PCR method was used for this purpose. Earlier reports have described specific primers to detect KIR alleles for genotyping and have also been utilized to detect mRNA-cDNA by various methods including quantitative real time PCR (Q-PCR) [5–7]. However, there are limitations to these methods and not all published primers amplify KIR cDNA using the Q-PCR method, especially if the amplified sequence is large (i.e., greater than 200–300bp). In this report, we developed a method to quantify KIR2DS2 and KIR2DS4 gene expression using previously cryo-preserved PBMCs, and to establish a baseline of aKIR gene expression in donor samples with corresponding genotype prior to HCT. Activating KIR expression was followed longitudinally at various times post-transplant and the relationship of aKIR expression to CMV reactivation and to GVHD was evaluated. In addition, CMV infection, but not GVHD, was associated with increased aKIR gene expression.
Materials and Methods
Study patients
Two hundred and eleven consecutive allogeneic HCT recipients, transplanted in the period 2001–2006, were followed for CMV reactivation at least for 1 year post-transplant [4]. From this study, cryo-preserved PBMCs samples from 134 HCT recipients, whose genotype was either KIR2DS2 or KIR2DS4 or both, were available for further analysis at various times post-HCT (days 40, 90, 120, 150 and 180). In addition, 81 donor samples were also available as part of the study, of which 55 were from donor – recipient pairs. Based on study eligibility, all HCT recipients were at risk for CMV reactivation as determined by CMV antibody seropositivity in either the donor or recipient or both. All subjects, donors and recipients, signed an informed consent for research approved by the City of Hope (COH) Institutional Review Board and the use of the left-over specimens for this study is covered under a COH IRB protocol. Table 1 describes the general demographics of the 134 study participants in terms of median age, transplant type, disease diagnosis, myeloablative vs. non-myeloablative conditioning, and GVHD prophylaxis.
Table 1.
Demographic status of the patient population
| Study cohort n = 134 | median (range ) or N (Percent) |
|---|---|
| Recipient Age at Transplant | 42.1 (18.8., 63.6) |
|
| |
| Donor Age at Transplant | 42.1 (18.0, 71.4) |
|
| |
| Donor Type | |
| Sibling Donor | 87 (65%) |
| Unrelated Donor | 47 (35%) |
|
| |
| Material Transplanted | |
| Bone Marrow Stem Cells | 24 (18%) |
| Peripheral Blood Stem Cells | 110 (82%) |
|
| |
| Diagnosis | |
| Lymphoid | 51 (38%) |
| Myeloid | 77 (57%) |
| Other | 6 (5%) |
|
| |
| Disease Status at Transplant | |
| 1st Complete Remission/Chronic Phase | 57 (43%) |
| Later CR/Later CP | 16 (12%) |
| Relapse | 22 (16%) |
| Induction Failure | 22 (16%) |
| MDS, AA, MM, MPD | 17 (13%) |
|
| |
| Conditioning | |
| Fludarabine/Melphalan | 45 (34%) |
| Myeloablative | 88 (66%) |
|
| |
| CMV serology | |
| missing | 1 |
| D−/R+ | 33 (25%) |
| D+/R− | 14 (10%) |
| D+/R+ | 86 (65%) |
|
| |
| Acute GVHD Grade | |
| 0-I | 53 (40%) |
| II-IV | 81 (60%) |
|
| |
| Chronic GVHD Grade | |
| None | 21 (16%) |
| Limited | 15 (11%) |
| Extensive | 98 (73%) |
AA: aplastic anemia; MM; multiple myeloma
CMV reactivation
CMV reactivation was monitored by DNA Q-PCR on plasma collected twice weekly up to 100 days post-HCT. CMV surveillance was continued at 1–2 week intervals in “high-risk” patients based on clinical management guidelines at City of Hope (COH). High-risk patients included those with persistent lymphopenia, grade 2–4 GVHD, or those requiring continued immunosuppression for anti-GVHD therapy. The Q-PCR was performed essentially as previously described using the CMV-gB DNA as amplification product [8] and had a lower limit of detection of 200 genome copies per milliliter plasma (gc/ml). In addition to Q-PCR testing on plasma, all patients had whole blood cultured for CMV infection on the same blood specimen using a shell vial culture method previously described [9]. In this study, CMV infection was defined as evidence showing CMV in any of three tests, namely, blood culture shell vial, plasma Q-PCR or CMV disease by histology. Time to CMV reactivation was defined as the day of first CMV positive Q-PCR (>200gc/ml), and the CMV infection was measured quantitatively by plasma DNA gc/ml.
KIR genotyping and expression
The method of Sun et al [10] was used to identify the 11 functional, non-framework genes (2DS1-5;3DS1;2DL1-3 and 2DL5;3DL1) and distinguish full-length from deleted alleles of KIR2DS4 in all subjects (see supplement figure S1). The KIR2DS2 and 2DS4 expression was done essentially according to Cooley et al [6, 11]. In brief, mRNA was processed from 5×106 PBMCs using RNeasy Mini kit (Qiagen, Valencia, CA), followed by DNAse treatment and first strand cDNA synthesis using Superscript II and oligo dT (Invitrogen). The RNA was degraded by the addition of Rnase H (Invitrogen). The cDNA equivalent of 125 ng of total RNA was used for each amplification reaction. The synthetic oligonucleotides (http://idtdna.com; Integrated DNA technologies, Coralville, IA) were designed with publicly available KIR sequences (Genbank; http://ncbi.nlm.nih.gov and IPD-KIR Sequence database; http://ebi.ac.uk/ipd/kir) and overlapped exons to ensure RNA sequence amplification. SYBR green was used for detection with the melting point dissociation curves as confirmation of specificity (see supplement figure S2). Actin amplification served as control (with a minimum Ct value of 30 or 1024 genome copies) and all the data presented here are expressed as copy number/1 x106 β-actin copies. The reactions were performed using the TaqMan Universal Master Mix and a 7900 Q-PCR Equipment (Applied Biosystems). The primers are as follows:
-
KIR2DS2: position: 198-454 NM_012312.2; size: 257bp [6]
FWD: (exon 3-198) -5’ TGCACAGAGAGGGGAAGTA 3’
REV: (exon4-454) -5’ CACGCTCTCTCCTGCCAA 3’
-
KIR2DS4: position 374-593 NM_012314.3; size 220bp
FWD: (exon 3-374) -5’ CAGTTGTCAGCTCCCAGTGA 3’
REV: (exon 4-593) -5’ CCTGGAATGTTCCGTTGATG 3’
-
B-ACTIN: position 317-504 NM_001101.3; size 188bp
FWD: (exon 3-317) -5’ ACTGGGACGACATGGAGAAA 3’
REV: (exon 4-504) -5’ TAGCACAGCCTGGATAGCAA 3’
The primers described here to amplify KIR2DS4 cDNA will not distinguish between the full length and the deletion mutant. The amplified sequences of KIR2DS2 and KIR2DS4 were verified by sequencing and the corresponding product was confirmed. The primers detected all 2DS2 and 2DS4 alleles except for 2DS2*00104, 2DS4*0040101 and 2DS4*013. Although these 3 alleles could not be analyzed for expression, they are a rare event and were not described by genotype in our HCT cohort.
Statistical analysis
The GraphPad Prism®5 software (GraphPad Software Inc., La Jolla CA) (www.graphpad.com) was used for the analysis of CMV DNA Q-PCR results. All other analyses were performed using the SAS® 9.25 (SAS Institute Inc., Cary, NC) and S-Plus® (Insightful Corp., Seattle, WA). The following tests were used: Mann-Whitney test for 2 samples, Kruskal-Wallis test for more than two samples, Wilcoxon signed-rank test for paired data. Contingency tables were analyzed using Fisher’s exact test. To evaluate whether the time to CMV reactivation was a factor in the expression of KIR2DS2 and KIR2DS4, Kaplan-Meier estimates were generated using the log-rank test. Logistic regression was performed and tested using Wald’s tests. All tests were two-sided and the cutoff for statistical significance was 0.05.
Results
Patient demographics
The material used in this study was cryopreserved PBMCs of the appropriate genotype collected from a previous study, as described in Material and Methods. The population demographics shown in Table 1 indicated that this subset of patients did not differ in the HCT parameters from the entire set previously described [4]. Specifically, the study population of 134 recipients had a median age of donor (42yr) and recipient (42yr), type of transplant: sibling 65%; unrelated 35%, and diagnoses: lymphoid 38%; myeloid 57%, similar in distribution to the full study cohort. The same was true for Donor/Recipient CMV-sero-positivity: D+/R+ 65%; D+/R− 10%; D−/R+ 25% and GVHD criteria: acute 0-I 40%; II-IV 60%. Of the 134 subjects, 98 (73%) became CMV viremic or had CMV-associated disease post-HCT as shown by PCR, blood culture or histopathology and 36 (27%) were not viremic.
KIR2DS2 and KIR2DS4 expression by mRNA Q-PCR in control samples
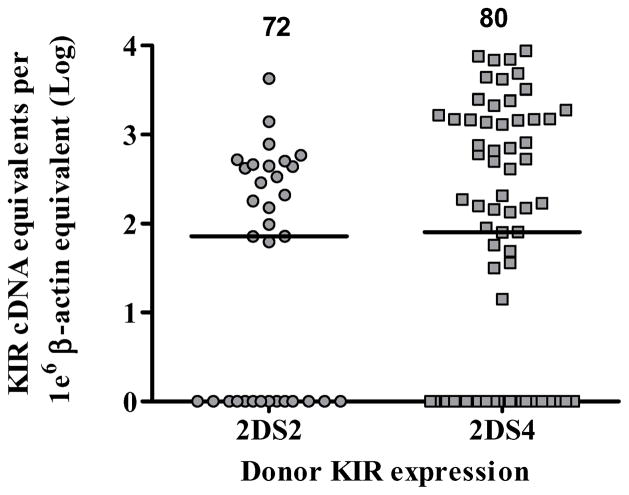
KIR expression in 81donor samples, collected after G-CSF treatment, ranged from 0–4.2×103 cDNA equivalent unit (e.u.) for KIR2DS2, and 0– 1.3×104 e.u. for KIR2DS4 with median values of 72 and 80, respectively (Figure 1). The KIR2DS4d mutant was identifiable by genotyping but not by specific mRNA expression, and therefore the overall expression of KIR2DS4, regardless of its allele is shown. The median expression in donor samples of both activating KIR genes were similar as were the percentage of null-expressers, 35% for KIR2DS2 and 38% for KIR2DS4. Of note, among the donors with both KIR2DS2 and 2DS4, dual KIR expression occurred in 50% of the subjects with no predominance of either gene. A baseline of KIR expression was thus established in the donor group, and this could be compared to KIR2DS2 and KIR2DS4 expression levels after transplantation.
Figure 1. KIR2DS2 and KIR2DS4 expression by mRNA-based Q-PCR in the donor.
KIR2DS2 and KIR2DS4 expression was reported only in the presence of the corresponding genotype. KIR2DS2 is represented in circles and KIR2DS4 in squares. The copy number for each subject was normalized to 1×106 β-actin copies and the median copy number is shown at the top of each group. The null-expressers are represented as log 100=1 on the y axis.
Since our hypothesis stated that aKIR expression has a role in protection for CMV reactivation, we examined whether the CMV serology of the donor as evidence of latent CMV infection, may have affected KIR expression. To answer this question, KIR2DS2 and 2DS4 expression were dichotomized into D- and D+ serology group. The median aKIR expression in D- and D+ cells were not statistically different by Mann-Whitney test suggesting that CMV serology of the donor did not ultimately affect KIR expression (results not shown).
KIR2DS2 and KIR2DS4 expression in Donor/Recipient pairs is elevated after HCT
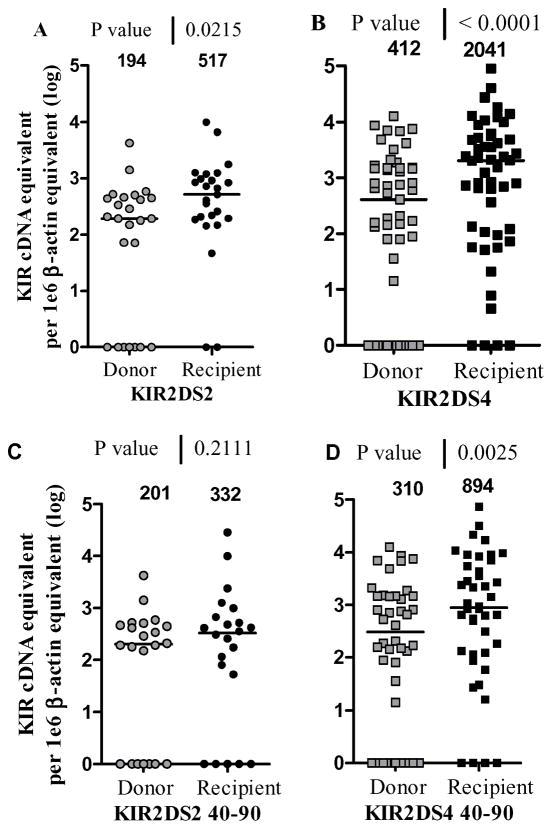
To determine whether the expression of KIR2DS2 and KIR2DS4 was affected after transplantation, the levels of aKIR were reported in donor–recipient pairs. Since full chimerism was reached in 95% of HCT recipients by day 30 post-HCT, it was the donor KIR profile that was analyzed for KIR expression in the recipient [12]. Each recipient was represented by mean equivalent units (e.u.) of multiple samples collected at the specific times of the study post-HCT (Figure 2). Overall, both KIR2DS2 (Figure 2A) and KIR2DS4 (Figure 2B) expression were significantly elevated after transplant. The median values in 25 KIR2DS2 pairs were 194 e.u. for donors and 517 e.u. in recipients (p= 0.02) and 412 e.u. for donors vs 2041 in recipients in 51 KIR2DS4 pairs (p<0.0001). Thus, Q-PCR results on mRNA purified from PBMCs shows that KIR2DS2 and 2DS4 levels were elevated after transplant. More importantly, KIR2DS4 levels were significantly higher at day 40–90 post-HCT when compared to the corresponding donor levels (median 310 e.u. for donors vs. 894 e.u. for recipients in 42 pairs with p value= 0.0025) (Figure 2D) but not with KIR2DS2 (Figure 2C).
Figure 2. KIR2DS2 and KIR2DS4 levels in Donor –Recipient pairs.
The activating KIR levels are reported for the donor before infusion (grey), and for its recipient post-HCT (black). The copy number for each subject was normalized to 1×106 β-actin copies and the median copy number is shown at the top of each group. A mean of all samples per recipient after HCT is shown in A and B and a mean of samples before day 90 post-HCT per recipient is shown in C and D. A) KIR2DS2 levels are shown for 25 D/R pairs and analyzed using the Wilcoxon paired rank test (p=0.02), B) KIR2DS4 levels are shown for 51 D/R pairs and analyzed using the Wilcoxon test (p<0.0001), C) KIR2DS2 levels are shown for 22 D/R pairs and analyzed using the Wilcoxon test (p=0.21), D) KIR2DS4 levels are shown for 42 D/R pairs and analyzed using the Wilcoxon test (p<0.0025). The null-expressers are represented as log 10 0=1 on the y axis.
Although 6/26 KIR2DS2 and 13/51 KIR2DS4 donors were expression negative prior to HCT, it was rare that in the recipient, there was no expression and so, KIR expression level in donor cells could not predict KIR expression levels in recipients. This is illustrated in Figure 2A for KIR2DS2 where the 2 recipient KIR null expressers (R−) were characterized by KIR D+/R− and D−/R− and KIR2DS4 results were characterized by 4 recipients KIR null expressers as follows: D−/R−, D+/R−, D−/R−, D−/R−. This D/R paired group showed very clearly that the 23–25% null expressers in donor cells have decreased to 8% null expressers in the recipient cells and emphasize the concept that transplantation activated KIR expression in HCT recipients.
KIR2DS2 and KIR2DS4 expression and CMV reactivation after HCT
To determine whether CMV reactivation detected after HCT affected KIR2DS2 and KIR2DS4 expression, HCT recipients were dichotomized as viremic (V) or non-viremic (NV) (Figure 3A) and the expression of KIR2DS2 was significantly upregulated in the viremic group, with median values of 117 e.u. for non-Viremic and 232 e.u. for Viremic subjects (MW test p=0.01). The upregulation was less pronounced for KIR2DS4 with median 386 e.u. for non-viremic and 769 e.u. for viremic subjects (p=0.06). However, if the HCT recipients were grouped according to donor genotype as shown in Figure 3B, KIR2DS4 full length expressed a significantly higher number of copies in the viremic group (p= 0.007) whereas heterozygotes for KIR2DS4 and KIR2DS4d mutant did not (p= 0.33) neither did the KIR2DS4d only (p=0.13). Of note, the latter group, consisting of the deletion mutant, expressed mRNA at extremely low levels, with medians that were more than 10 fold less than the expressed KIR2DS4. These results emphasize the importance for detecting deletion mutant alleles that do not express KIR receptors. Of note, though not statistically significant, transplant recipients of dual 2DS2 and 2DS4 donor genotype also exhibited an upregulation of expression in the Viremic group (Figure 3C). CMV disease occurs in 10 % of CMV reactivation in our institution and the question was whether KIR expression was different between patients who developed CMV disease from those who did not. The results are shown in table 2 for KIR2DS2 (3 CMV disease and 49 viremic patients) and KIR2DS4 (4 CMV disease and 55 viremic patients). Overall, the median KIR expression was lower in CMV disease patients and significantly lower with KIR2DS2 expression (p=0.0105).
Figure 3. KIR2DS2 and KIR2DS4 expression by mRNA-based Q-PCR in HCT subjects.
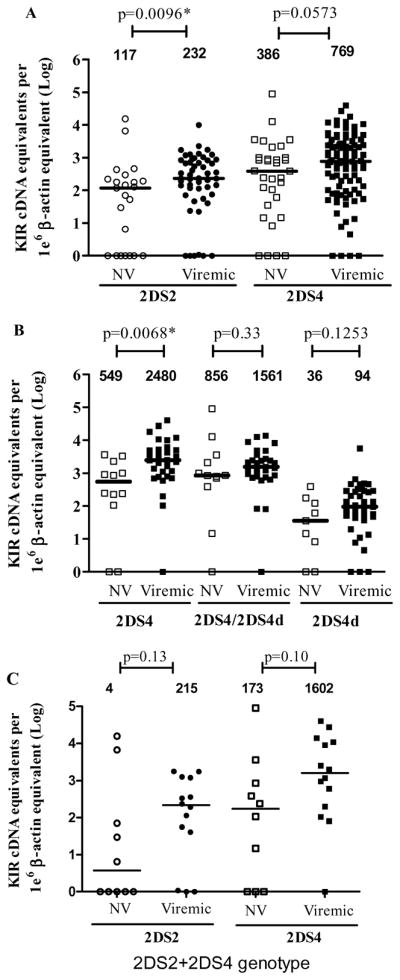
The samples from non-viremic (NV) subjects are in clear symbol and samples from viremic subjects in black. KIR2DS2 is represented in circles and KIR2DS4 in squares. A mean value was reported per patient and the KIR expression values were reported only if the corresponding donor KIR genotype was present. The copy number for each subject was normalized to 1×106 β-actin copies and the median copy number is shown at the top of each group. A Mann-Whitney (MW) test was performed to establish p values between the NV and Viremic groups. Panel A) shows the overall expression of either KIR 2DS2 or KIR2DS4 in both groups NV and V; B) differentiates between the expression of KIR2DS4 and the deletion mutant KIR2DS4d and C) shows the results for recipients receiving both genotypes together from donor cells. The null-expressers are represented as log 100=1 on the y axis.
Table 2.
KIR expression comparing CMV Disease vs viremic patients
| 2DS2 | 2DS4 | |||||
|---|---|---|---|---|---|---|
| CMV disease | Viremic | M.W. test* | CMV disease | Viremic | M.W. test* | |
| number of patients | 3 | 49 | 4 | 55 | ||
| Mean expression | 7.97 | 708.14 | 2136.91 | 4530.6 | ||
| Median expression | 0 | 306.96 | p=0.0105 (S) | 1404.99 | 2164.41 | p=0.5633 (NS) |
| Standard error | 4.6 | 29.55 | 501.64 | 127.23 | ||
M.W. test: Mann-Whitney test
Time course of KIR expression post-HCT
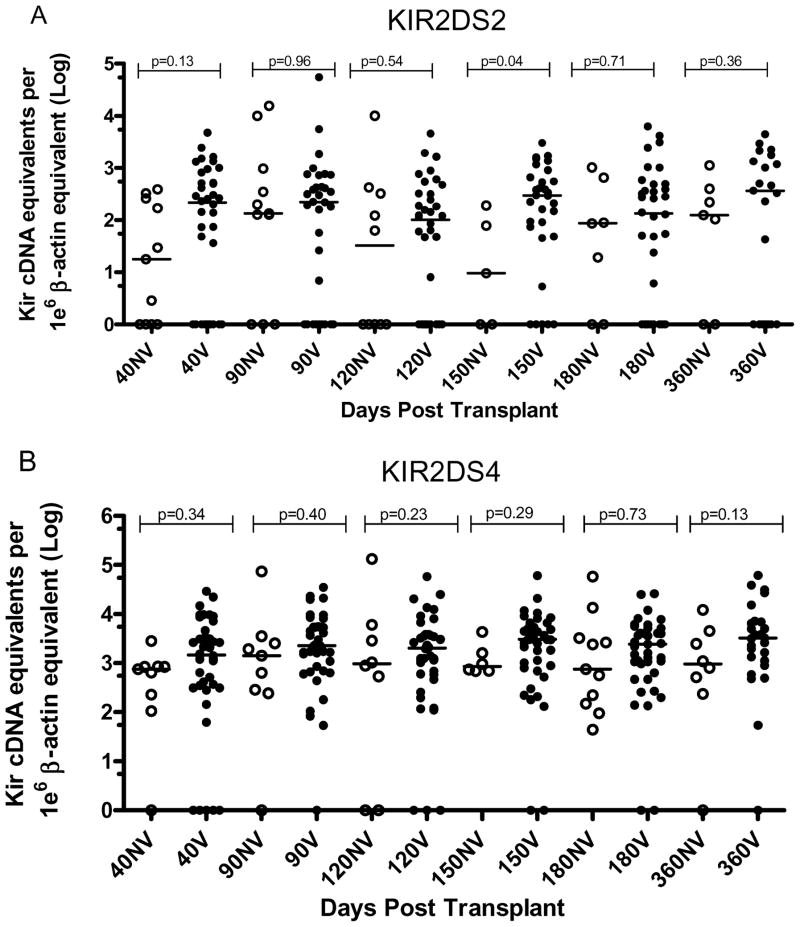
Blood samples were available at day 40, 90, 120, 150, 180 and 360 days post-HCT. This sampling schedule allowed us to analyze the impact of time on aKIR expression after HCT. Kruskal Wallis test and Dunn’s multiple comparison tests were performed on each data set, 2DS2 NV vs 2DS2 Viremic (Figure 4A), and 2DS4 NV vs 2DS4 Viremic (Figure 4B). The recipients with the KIR2DS4d mutant genotype only were not included in the analysis. The p values showed no significant differences according to time among the group of expressers within each subgroup.
Figure 4. Time course of KIR expression post-HCT in non viremic and viremic subjects.
Cryopreserved PBLs were available for days 40, 90, 120, 150, 180, and 360 post-transplant. The presence of at least one full length genotype KIR2DS4 allele was a condition to be included in panel B. A) shows the results for KIR2DS2 expression in non viremic subjects vs viremic subjects (Mann –Whitney test p values are shown); B) describes the expression of KIR2DS4 in non viremic subjects vs viremic subjects (Mann-Whitney test). The median values are depicted above each column. The null-expressers are represented as log 100=1 on the y axis.
CMV reactivation usually occurred between day 40 and 90 post-HCT. To explore whether a difference of KIR expression could be seen non-viremic vs viremic patients during that critical period, KIR2DS2 NV expression was compared to 2DS2 Viremic at day 40–90 and no statistical difference was detected. The same was done for KIR2DS4 expression. Thus, when expression was analyzed without regard to the time of CMV reactivation, no difference was detected between the non-viremic and the viremic KIR expression post-HCT.
KIR expression status before CMV reactivation
To establish whether KIR2DS2 or KIR2DS4 expression before CMV reactivation would influence CMV outcome, a cumulative incidence curve for CMV reactivation was generated for expressers or null-expressers for each aKIR as shown in Figure 5A and 5B. The following criteria were followed for this analysis: since KIR expression data was available only on or after day 40 post-HCT, no CMV reactivation event occurring before day 40 was included here; however, KIR expression data at day 40 with CMV reactivation at the same time were included. CMV reactivation occurred in 42% of KIR2DS2 null-expressers vs 54% expressers and 57% of KIR2DS4 null–expressers vs 51% expressers. The log-rank test p values shows that for HCT subjects who had CMV reactivation, there was no difference whether the specific aKIR were expressed before reactivation or not (see figures 5A and 5B). However, recipients with both 2DS2 and 2DS4 expression (see Figure 5C) shows that CMV reactivation occurred in only 22% of the patients (p=0.04).
Figure 5. Effect of KIR expression status before CMV reactivation.
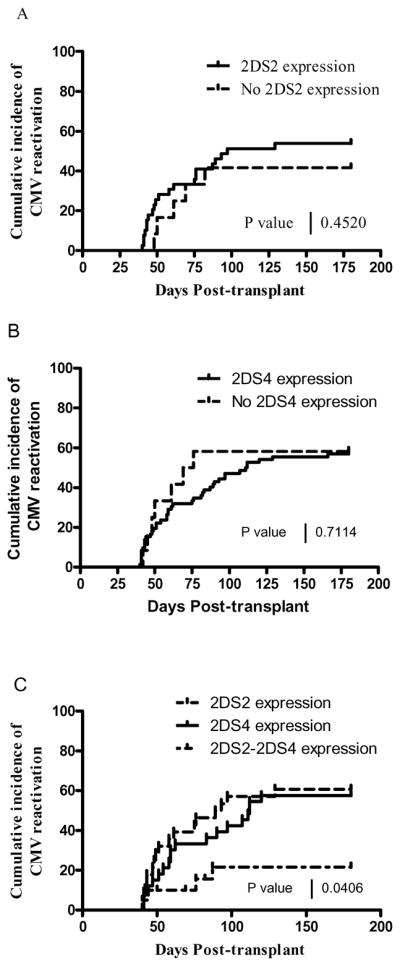
A) shows the percent cumulative CMV incidence in the presence or absence of KIR2DS2 expression before CMV reactivation (log-rank test p=0.45) and B) shows the same plot applied to KIR2DS4 (log-rank test p= 0.51). C) shows only the presence of expression ( greater than 0) for either 2DS2, or 2DS4, or 2DS2+2DS4 before CMV reactivation (log-rank test p=0.04).
HCT variables affecting KIR2DS2 and 2DS4 expression
Having determined that KIR2DS2/4 expression was upregulated in CMV viremic subjects, the question was whether this was related to other factors of allotransplantation variables such as donor type, transplant source, diagnosis, disease status at transplant, conditioning regimen, CMV serology (Donor/Recipients), acute and chronic GVHD. As shown in Table 2 the median expression in recipients of matched unrelated donors was significantly lower than in recipients of sibling donor for both KIR2DS2 (p=0.0004) and KIR2DS4 (p=0.003). In addition, stem cells collected from the bone marrow were associated with decreased median expression of KIR2DS2 (p=0.04) and 2DS4 (p=0.03). Bone marrow contains pluripotent stem cells that give rise to all classes of blood cells and the low levels of NK cells could explain the detected low level of aKIR expression. In addition, in this cohort, bone marrow as a source of stem cells is found more often as a transplant source in the unrelated matched donor (33%) compared to sibling donors (9%) which explains the association of low level of KIR expression with both BM and unrelated matched transplant (p=0.0005). Also of note, neither the acute GVHD grade nor the chronic GVHD grade had a significant effect on KIR expression.
Multivariable analysis using CMV viremia as outcome
The sample population described here were known strictly by genotype 2DS2 (74 subjects) and/or 2DS4 (84 subjects), and the model analyzed each genotype independently with a baseline set at zero expression for each KIR type. As shown in Table 3, the donor type (for KIR2DS2 p=0.0004; for KIR2DS4 p=0.0002) and transplant source (for KIR2DS2 p=0.04; for KIR2DS4 p= 0.03) affected KIR expression but aGVHD had no effect on KIR expression. However, since aGVHD grade is an independent predictor of CMV viremia, we included all 3 variables into a multivariable logistic regression shown in Table 4.
Table 3.
Comparison of levels of expression of KIR by predictor variable level
| Predictor variable | KIR 2DS2 Expression a | KIR 2DS4 Expression b | ||
|---|---|---|---|---|
| Median (range) | P* | Median (range) | P* | |
| Donor Type | ||||
| Sibling Donor | 346 (0, 15624) | 0.0004 | 2302 (0, 89328) | 0.0002 |
| Unrelated Donor | 95 (0, 701) | 769 (0, 8304) | ||
|
| ||||
| Transplant source | ||||
| Bone Marrow | 94.5 (0, 1038) | 0.04 | 834 (15, 4863) | 0.03 |
| Peripheral Blood Stem Cells | 232 (0, 15624) | 2053.5 (0, 89328) | ||
|
| ||||
| Diagnosis | ||||
| Lymphoid | 178 (0, 2265) | 0.53 | 1561 (0, 15107) | 0.91 |
| Myeloid | 228 (0, 15624) | 1488 (0, 89328) | ||
| Other | 1 (0, 1038) | 910 (104, 8304) | ||
|
| ||||
| Disease Status at Transplant | ||||
| 1st Complete Remission/CP | 188.5 (0, 15624) | 0.96 | 2066 (0, 15107) | 0.46 |
| Later CR/Later CP | 224 (6, 1254) | 863 (15, 2725) | ||
| Relapse | 232 (0, 2265) | 2209.5 (81, 5224) | ||
| Induction Failure | 178 (0, 1769) | 1235 (0, 27388) | ||
| MDS, AA, MM, MPD | 194 (0, 6648) | 2041 (104, 89328) | ||
|
| ||||
| Conditioning | ||||
| Fludarabine/Melphalan | 225.5 (0, 2265) | 0.75 | 2033 (0, 39860) | 0.83 |
| Myeloablative | 214.5 (0, 15624) | 1383 (0, 89328) | ||
|
| ||||
| CMV Serology | ||||
| D−/R+ | 76 (0, 1758) | 0.03 | 1452 (0, 8304) | 0.75 |
| D+/R− | 128 (0, 15624) | 2079 (0, 5005) | ||
| D+/R+ | 300 (0, 10023) | 1449 (0, 89328) | ||
|
| ||||
| Acute GVHD Grade | ||||
| 0-I | 188.5 (1758) | 0.17 | 1110 (0, 39860) | 0.55 |
| II-IV | 268.5 (0, 15624) | 1561 (0, 89328) | ||
|
| ||||
| Chronic GVHD Grade | ||||
| None | 321 (0, 1769) | 0.53 | 1839 (0, 27388) | 0.83 |
| Limited | 300 (0, 1254) | 1226 (0, 5224) | ||
| Extensive | 194 (0, 15624) | 2033 (0, 89328) | ||
Kruskal-Wallis Test;
KIR2DS2 genotype;
KIR2DS4 genotype.
Table 4.
Multivariable Logistic Regressions—Probability Model CMV status = Viremic
| Predictor | N | Odds Ratio (95% CI) | P-value | N |
|---|---|---|---|---|
| KIR 2DS2 Expression a | 0.04* | 74 | ||
| Zero, | 10 | (baseline) | ||
| Greater Than Zero | 64 | 7.16 (1.51 – 33.9) | 0.01 | |
| Donor Type | ||||
| Unrelated matched donor | 28 | (baseline) | ||
| Sibling Donor | 46 | 1.00 (0.29 – 3.48) | 1.00 | |
| Transplant source | ||||
| BM | 14 | (baseline) | ||
| PBSC | 60 | 3.89 (0.96 – 15.8) | 0.06 | |
| Acute GVHD | ||||
| O, I | 28 | (baseline) | 0.27 | |
| II, III, IV | 46 | 1.93 (0.61 – 6.12) | ||
|
| ||||
| KIR 2DS4 Expression b | 0.009* | 84 | ||
| Zero, | 5 | (baseline) | ||
| Greater Than Zero | 79 | 6.69 (0.88 – 51.0) | 0.07 | |
| Donor Type | ||||
| Unrelated matched donor | 29 | (baseline) | 1.00 | |
| Sibling Donor | 55 | 1.00 (0.29 – 3.46) | ||
| Transplant source | ||||
| BM | 14 | (baseline) | 0.02 | |
| PBSC | 70 | 5.08 (1.27 – 20.3) | ||
| Acute GVHD | ||||
| O, I | 31 | (baseline) | 0.009 | |
| II, III, IV | 53 | 4.61 (1.46 – 14.6) | ||
Goodness of Fit of the model
KIR2DS2 genotype ;
KIR2DS4 genotype.
The Odds Ratio for a patient with KIR2DS2 expression >0 was 7 times more likely to occur in viremic patients --O.R. 7.16 (1.51 – 33.9). Adding to this logistic regression, the donor type --O.R. 1.00(0.29 – 3.48), transplant source--O.R. 3.89 (0.96 – 15.8) and aGVHD --O.R. 1.93 (0.61 – 6.12), the goodness of fit of the model was still significant (p=0.04).
Similarly for KIR2DS4 expression, the Odds Ratio for expression to be greater than 0 in viremic subjects was 6.69 (0.88 – 51.0) with a significance of p=0.07. If donor type --O.R.1.00 (0.29 – 3.46)--, transplant source --O.R. 5.08 (1.27 – 20.3) and aGVHD --O.R. 4.61 (1.46 – 14.6) were included in the model, the goodness of fit was p=0.009.
KIR expression and HLA ligand
Activating KIR expression could be modulated by the presence of HLA specific ligand either in the donor or the recipient. Whereas KIR2DS2 binds with HLA C1 with weak affinity, KIR2DS4 can bind to multiple ligands with various affinities such as HLA *A11:01, HLA *A11:02, *C02, *C05, *C14, and *C16 [13]KIR 2DS2 ligand C1/C1 was present in 16% of donor and in 22% of recipient but KIR2DS2 ligand C1/C2 was present in 41% of donors and in 32% of recipients. HLA A*11:01 was present in 27% of KIR2DS4+ donors and in 18% of the recipients. The other HLA C were pooled together because they were scarce (8% of donors and 17% of recipients). The statistical analysis of proportions of subjects expressing KIR in the presence or absence of ligands show that there is no difference by Fisher’s exact test, even if subgroups are dichotomized between non-viremic or viremic patients (see supplementary Table 1). Thus, activating KIR expression does not seem to be driven by the presence of ligand but there are too few patient numbers to validate the power of the statistical test.
Discussion
The presence of activating KIR genes in the donor has been shown to have an important role in protection from such events as poor graft function [14], leukemic relapse [15–16], and CMV reactivation [4]. In the report showing a protective effect on CMV infection in HCT recipients having a donor with both a KIR2DS2 and KIR2DS4 genotype, it was noted that nearly half of those with this protective genotype still had CMV reactivation. Thus, it was hypothesized that failure of protection would be explained by altered expression of these KIR genes. This report focused on expression of KIR2DS2 and KIR2DS4, and asked whether their expression correlated with protection from CMV reactivation. KIR expression was assessed by Q-PCR of purified mRNA taken at intervals during the major risk period for CMV reactivation after HCT. We chose Q-PCR because it discriminated KIR2DS2 and 2DL2, as well as allowing for use of a quantitative approach to statistical analysis. Expression data was normalized to β-actin mRNA equivalent units. Using this quantitative method, the overall expression of the 2 KIR genes was assessed and compared to clinical events after transplant. There was no attempt to discriminate between the specific contribution of NK cells or T-cells and therefore KIR expression could come from both cell types. Due to sample limitation, we were unable to report NK and T cell counts for each sample and the upregulation of KIR expression could be the result of a simple increase in NK or T cells number. Other framework KIRs like 2DL4 and 3DL2 could also have been used as reference genes to evaluate specific KIR upregulation and these deserve another study.
The data demonstrate that in donor cells, the expression of KIR2DS2 and KIR2DS4 varies among genotype-positive healthy individuals (the donors), with some expressers and some non-expressers. Under these donor conditions, expression of these genes was at low levels (median 72 and 80 e.u. respectively), and about 35% of these healthy donors were non-expressers. The CMV serology of the donor had no impact on KIR expression. However, once transplanted, KIR expression of 2DS2 and 2DS4 was upregulated in HCT recipients. In paired D/R samples from matched related donor (MRD) allotransplants, the upregulation could be shown in the period d40 to d90 post-HCT for KIR2DS4 (p= 0.0025) (see Figure 2), the earliest time period examined. Of particular note, the level of expression in the recipients of grafts from unrelated matched donors was significantly lower than in MRD recipients (see Table 2). This is consistent with the improved outcome of MRD transplant recipients [17], and it suggests that receptor-ligand interactions deserve further study to delineate these differences. In addition, BM as transplant source was found at a higher rate in unrelated matched donors of (33% vs 9% for PBMCs) and this could account for low KIR expression since it has been shown that BM contains 20 times fewer NK cells in the graft [18].
We then asked whether this increased KIR expression in HCT recipients was associated with specific clinical events post HCT such as CMV reactivation. Figure 3 shows the plot of mean KIR expression of KIR2DS2 and 2DS4 per subject in the non-viremic and the CMV viremic groups. KIR levels were significantly higher in the viremic group for both KIRs. The KIR2DS4d mutant, known not to be expressed on the cell surface, had much lower levels of mRNA transcripts (median: 36 e.u. in NV group) than the full length (median: 549 e.u. in NV group). To determine whether the CMV reactivation was related to non-expression of these KIR genes prior to reactivation, we examined cumulative mean expression levels as well as expression at multiple time points prior to and after CMV reactivation. As shown in Figure 3, the mean KIR expression levels at all time points for each patient were significantly higher in the viremic group. However, when the time course of expression post-HCT was analyzed, we could see no statistical differences between the viremic and the non-viremic groups. It is likely that this is due to the variable time of CMV reactivation post-HCT. Of note, CMV infection has been associated with higher levels of NK cells and this could well explain in part the increase in KIR expression in those with infection [19].
We hypothesized that if aKIR expression affected CMV reactivation, we should see a difference in the cumulative incidence of CMV infection in the expresser vs non-expresser group. However, as shown in Figure 5, there was no difference in the cumulative incidence of CMV reactivation in these two groups (42% vs 54% for KIR2DS2 null-expressers and expressers, respectively, and 57% vs 51% for KIR2DS4 null-expressers and expressers, respectively). The ratio of null-expressers for KIR2DS2 was 5/21(19%) and for KIR2DS4, 4/25 (14%), and, as previously shown (Figure 3), the non-viremic patients actually had a lower expression of aKIR. However, CMV reactivation occurred in only 22% of the patients who expressed both KIR2DS2 and KIR2DS4 before CMV onset. Thus, we suggest that multiple aKIR expression such as KIR2DS2 and KIR2DS4, may protect from CMV reactivation strengthening our earlier report based on genotyping [4].
The question then was whether the upregulation of aKIR expression was really a response to cofactors which associate with CMV, for example GVHD. To check whether this or other transplant factors were responsible for KIR upregulation, KIR expression levels were analyzed against various transplant variables as shown in Table 2. The donor type, sibling vs unrelated matched donors, and transplant source, BM vs peripheral blood progenitor cells were variables that significantly affected the KIR expression levels. Both variables have in common the fact that BM does not contain mature NK and T cells and is frequently the transplant source in unrelated matched transplant. In contrast, no significant difference was found between expression levels of KIR and severity of aGVHD. In a multivariable analysis, in which the above mentioned transplant predictors of CMV viremia were included (see Table 4), KIR2DS2 expression greater than 0 was still predictive of viremia as an outcome, and the same was true for KIR2DS4. It is known that aGVHD (II-IV) grade is significantly associated with CMV viremia [4]; nevertheless, aKIR expression was associated only with CMV infection and not with aGVHD per se. Thus, it is concluded that several factors can influence KIR gene expression post-HCT, and that acute GVHD has little influence, but once CMV infection occurs there is a strong stimulus for KIR2DS2 and 2DS4 expression.
The presence of HLA ligand specific to KIR has been shown to drive KIR expression into a “missing self” recognition and lead to proliferation of NK cells to eliminate stressed, virally infected or transformed cells. Activating and inhibitory KIRs are the results of an evolutionary repertoire of receptors that bind both host and pathogen-encoded ligands [20]. The ligand for KIR2DS2 has been shown to be HLA C1 [21]and for KIR2DS4, a variety of ligand binding with various affinities have been described by Graef et al [13]. They include HLA *A11:01, *A11:02, *C16, 14, 05, and 02. It is not clear whether in our small cohort of patients the presence of ligands specific to KIR2DS2 and KIR2DS4 were a key factor responsible for their respective KIR expression. Interestingly, there are increased mRNA levels after transplant and even higher levels in viremic subjects. Early reports have shown that full effector function of NK cells requires triggering of their activating receptors via stress-induced or virus-encoded ligand on target cells but the activating NK cell receptors and ligands responsible for mediating this effect remains elusive [22].
Murine studies [23–24] have shown that if mature NK cells from wild-type are transferred into a host with a different MHC class I environment, the cells can be reprogrammed to their new environment. This may explain the increased expression of activating KIR after transplant seen here. Mature NK cells can be re-educated as their new environment dictates, the new environment being allo-transplantation and /or CMV reactivation. Of note, MRD transplant typically do encode for the same HLA type in donor and recipient but, it is possible that minor histocompatibility antigens or other HLA (e.g. nonclassical HLA-G-E) could serve as ligand that could influence this enhancement of expression.
Finally, the question is what specific CMV-associated factors might influence KIR gene expression? KIR genes are known to be regulated by epigenetic factors [25–27], and it is possible that either viral or host factors have some effect. We show here that at steady-state levels, normal individuals have varying levels of non-expression, and this should be explored in more detail by analysis of known epigenetic markers. The promoter regions for KIR2DS2 and KIR2DS4 are incompletely understood, but it is known that coding regions for these genes (chromosome 19) are highly methylated [25–28]. In addition, a recent report by Cichocki et al suggests that it is the antisense transcripts that mediate KIR transcriptional silencing through a novel PIWI-like 28 base small RNA [29]. It would be important to determine if variations in expression in our model of CMV post-HCT are due to such cellular events. In our study design, early time-points post-HCT were not fully explored, and it is possible that the factors present early in cell differentiation post-transplant could significantly affect KIR gene expression and alter CMV reactivation.
Supplementary Material
Acknowledgments
This work was supported by US Public Health Service grant no. RO1 AI58148 and R01 CA145207 to JAZ; 5 P30 CA 33572-27 to Friedman; and M01-RR00043-38 to the General Clinical Research Center at City of Hope.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial disclosure: The authors have nothing to disclose.
References
- 1.Chen C, Busson M, Rocha V, et al. Activating KIR genes are associated with CMV reactivation and survival after non-T-cell depleted HLA-identical sibling bone marrow transplantation for malignant disorders. Bone Marrow Transplant. 2006;38:437–444. doi: 10.1038/sj.bmt.1705468. [DOI] [PubMed] [Google Scholar]
- 2.Cook M, Briggs D, Craddock C, et al. Donor KIR genotype has a major influence on the rate of cytomegalovirus reactivation following T-cell replete stem cell transplantation. Blood. 2006;107:1230–1232. doi: 10.1182/blood-2005-03-1039. [DOI] [PubMed] [Google Scholar]
- 3.Stern M, Elsasser H, Honger G, et al. The number of activating KIR genes inversely correlates with the rate of CMV infection/reactivation in kidney transplant recipients. Am J Transplant. 2008;8:1312–1317. doi: 10.1111/j.1600-6143.2008.02242.x. [DOI] [PubMed] [Google Scholar]
- 4.Zaia JA, Sun JY, Gallez-Hawkins GM, et al. The effect of single and combined activating killer immunoglobulin-like receptor genotypes on cytomegalovirus infection and immunity after hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2009;15:315–325. doi: 10.1016/j.bbmt.2008.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uhrberg M, Valiante NM, Shum BP, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 6.Cooley S, Xiao F, Pitt M, et al. A subpopulation of human peripheral blood NK cells that lacks inhibitory receptors for self-MHC is developmentally immature. Blood. 2007;110:578–586. doi: 10.1182/blood-2006-07-036228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yawata M, Yawata N, McQueen KL, et al. Predominance of group A KIR haplotypes in Japanese associated with diverse NK cell repertoires of KIR expression. Immunogenetics. 2002;54:543–550. doi: 10.1007/s00251-002-0497-x. [DOI] [PubMed] [Google Scholar]
- 8.Gallez-Hawkins G, Thao L, Lacey SF, et al. Cytomegalovirus immune reconstitution occurs in recipients of allogeneic hematopoietic cell transplants irrespective of detectable cytomegalovirus infection. Biol Blood Marrow Transplant. 2005;11:890–902. doi: 10.1016/j.bbmt.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Gleaves CA, Smith TF, Shuster EA, Pearson GR. Comparison of standard tube and shell vial cell culture techniques for the detection of cytomegalovirus in clinical specimens. J Clin Microbiol. 1985;21:217–221. doi: 10.1128/jcm.21.2.217-221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun JY, Gaidulis L, Miller MM, et al. Development of a multiplex PCR-SSP method for Killer-cell immunoglobulin-like receptor genotyping. Tissue Antigens. 2004;64:462–468. doi: 10.1111/j.1399-0039.2004.00303.x. [DOI] [PubMed] [Google Scholar]
- 11.Leung W, Iyengar R, Triplett B, et al. Comparison of killer Ig-like receptor genotyping and phenotyping for selection of allogeneic blood stem cell donors. J Immunol. 2005;174:6540–6545. doi: 10.4049/jimmunol.174.10.6540. [DOI] [PubMed] [Google Scholar]
- 12.Toor A, Rodriguez T, Bauml M, et al. Feasibility of conditioning with thymoglobulin and reduced intensity TBI to reduce acute GVHD in recipients of allogeneic SCT. Bone Marrow Transplant. 2008;42:723–731. doi: 10.1038/bmt.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graef T, Moesta AK, Norman PJ, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med. 2009;206:2557–2572. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cirocco RE, Mathew JM, Burke GW, 3rd, Esquenazi V, Miller J. Killer cell immunoglobulin-like receptor polymorphisms in HLA-identical kidney transplant recipients: lack of 2DL2 and 2DS2 may be associated with poor graft function. Tissue Antigens. 2007;69 (Suppl 1):123–124. doi: 10.1111/j.1399-0039.2006.76211.x. [DOI] [PubMed] [Google Scholar]
- 15.Verheyden S, Schots R, Duquet W, Demanet C. A defined donor activating natural killer cell receptor genotype protects against leukemic relapse after related HLA-identical hematopoietic stem cell transplantation. Leukemia. 2005;19:1446–1451. doi: 10.1038/sj.leu.2403839. [DOI] [PubMed] [Google Scholar]
- 16.Kroger N, Binder T, Zabelina T, et al. Low number of donor activating killer immunoglobulin-like receptors (KIR) genes but not KIR-ligand mismatch prevents relapse and improves disease-free survival in leukemia patients after in vivo T-cell depleted unrelated stem cell transplantation. Transplantation. 2006;82:1024–1030. doi: 10.1097/01.tp.0000235859.24513.43. [DOI] [PubMed] [Google Scholar]
- 17.Arora M, Weisdorf DJ, Spellman SR, et al. HLA-identical sibling compared with 8/8 matched and mismatched unrelated donor bone marrow transplant for chronic phase chronic myeloid leukemia. J Clin Oncol. 2009;27:1644–1652. doi: 10.1200/JCO.2008.18.7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dreger P, Haferlach T, Eckstein V, et al. G-CSF-mobilized peripheral blood progenitor cells for allogeneic transplantation: safety, kinetics of mobilization, and composition of the graft. Br J Haematol. 1994;87:609–613. doi: 10.1111/j.1365-2141.1994.tb08321.x. [DOI] [PubMed] [Google Scholar]
- 19.Guma M, Angulo A, Lopez-Botet M. NK cell receptors involved in the response to human cytomegalovirus infection. Curr Top Microbiol Immunol. 2006;298:207–223. doi: 10.1007/3-540-27743-9_11. [DOI] [PubMed] [Google Scholar]
- 20.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 21.Cook MA, Milligan DW, Fegan CD, et al. The impact of donor KIR and patient HLA-C genotypes on outcome following HLA-identical sibling hematopoietic stem cell transplantation for myeloid leukemia. Blood. 2004;103:1521–1526. doi: 10.1182/blood-2003-02-0438. [DOI] [PubMed] [Google Scholar]
- 22.Sun JC. Re-educating natural killer cells. J Exp Med. 2010;207:2049–2052. doi: 10.1084/jem.20101748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joncker NT, Shifrin N, Delebecque F, Raulet DH. Mature natural killer cells reset their responsiveness when exposed to an altered MHC environment. J Exp Med. 2010;207:2065–2072. doi: 10.1084/jem.20100570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elliott JM, Wahle JA, Yokoyama WM. MHC class I-deficient natural killer cells acquire a licensed phenotype after transfer into an MHC class I-sufficient environment. J Exp Med. 2010;207:2073–2079. doi: 10.1084/jem.20100986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li G, Yu M, Weyand CM, Goronzy JJ. Epigenetic regulation of killer immunoglobulin-like receptor expression in T cells. Blood. 2009;114:3422–3430. doi: 10.1182/blood-2009-01-200170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan HW, Miller JS, Moore MB, Lutz CT. Epigenetic control of highly homologous killer Ig-like receptor gene alleles. J Immunol. 2005;175:5966–5974. doi: 10.4049/jimmunol.175.9.5966. [DOI] [PubMed] [Google Scholar]
- 27.Santourlidis S, Trompeter HI, Weinhold S, et al. Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J Immunol. 2002;169:4253–4261. doi: 10.4049/jimmunol.169.8.4253. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Kuick R, Hanash S, Richardson B. DNA methylation inhibition increases T cell KIR expression through effects on both promoter methylation and transcription factors. Clin Immunol. 2009;130:213–224. doi: 10.1016/j.clim.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cichocki F, Lenvik T, Sharma N, et al. Cutting edge: KIR antisense transcripts are processed into a 28-base PIWI-like RNA in human NK cells. J Immunol. 2010;185:2009–2012. doi: 10.4049/jimmunol.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.