Background: Neisseria meningitidis NST catalyzes the transfer of sialic acid to the terminus of surface LOS.
Results: We present NST crystallographic and mechanistic data.
Conclusion: NST exhibits a novel homodimeric, composite active site, and lipid binding channel not observed in any other glycosyltransferase to date.
Significance: This work improves our understanding of lipopolysaccharide sialylation with the first crystallographic structure of a CAZY family-52 glycosyltransferase.
Keywords: Bacteria, Enzyme Structure, Kinetics, Oligosaccharide, X-ray Crystallography, Lipooligosaccharide, Neisseria meningitidis, Sialyltransferase
Abstract
The first x-ray crystallographic structure of a CAZY family-52 glycosyltransferase, that of the membrane associated α2,3/α2,6 lipooligosaccharide sialyltransferase from Neisseria meningitidis serotype L1 (NST), has been solved to 1.95 Å resolution. The structure of NST adopts a GT-B-fold common with other glycosyltransferase (GT) families but exhibits a novel domain swap of the N-terminal 130 residues to create a functional homodimeric form not observed in any other class to date. The domain swap is mediated at the structural level by a loop-helix-loop extension between residues Leu-108 and Met-130 (we term the swapping module) and a unique lipid-binding domain. NST catalyzes the creation of α2,3- or 2,6-linked oligosaccharide products from a CMP-sialic acid (Neu5Ac) donor and galactosyl-containing acceptor sugars. Our structures of NST bound to the non-hydrolyzable substrate analog CMP-3F(axial)-Neu5Ac show that the swapping module from one monomer of NST mediates the binding of the donor sugar in a composite active site formed at the dimeric interface. Kinetic analysis of designed point mutations observed in the CMP-3F(axial)-Neu5Ac binding site suggests potential roles of a requisite general base (Asp-258) and general acid (His-280) in the NST catalytic mechanism. A long hydrophobic tunnel adjacent to the dimer interface in each of the two monomers contains electron density for two extended linear molecules that likely belong to either the two fatty acyl chains of a diglyceride lipid or the two polyethylene glycol groups of the detergent Triton X-100. In this work, Triton X-100 maintains the activity and increases the solubility of NST during purification and is critical to the formation of ordered crystals. Together, the mechanistic implications of the NST structure provide insight into lipooligosaccharide sialylation with respect to the association of substrates and the essential membrane-anchored nature of NST on the bacterial surface.
Introduction
Sialic acid, commonly found at the terminal position of glycoconjugates (i.e. glycolipid and glycoproteins), activates a multitude of biological functions through specific recognition of carbohydrate receptors in humans (1). Sialic acid is a key player in the human immune system, including a role with Factor H in directing the alternative pathway of complement against foreign pathogens (2, 3), a role with selectin in guiding leukocytes to the site of inflammation (4), and a role with specific interleukins in regulating the onset of the immune response (5). However, many pathogens have evolved to take advantage of host surface-exposed sialic acids, and in bacterial pathogens such as Neisseria meningitidis this is evident in an invasion mechanism involving mimicry of the polysialic acid structure on host neural cell adhesion molecules by the capsular polysialic acid, which effectively camouflages the bacteria from the human immune system (6). Second, the structural similarity between the sialylated epitopes on host gangliosides (GM1, Gal(β1,3)GalNac(β1,4)[NeuAc(α2,3)]Gal(β1,4)Glc-(β1,1)ceramide) and bacterial lipooligosaccharide (LOS)2 can lead to development of the autoimmune neurodegenerative disorders Guillain-Barré and Miller Fisher syndromes, respectively (7, 8).
N. meningitidis is a neuroinvasive human pathogen causing meningitis and sepsis in infected patients. Members of the Neisseria genus may possess two antigenic structures on the outer membrane surface that include the extracellular capsule and endotoxic LOS, both of which can effect resistance to phagocytotic killing and suppression of the human immune response (9). For encapsulated Neisseria meningococci, the capsular polysaccharide plays a dominant role in effecting virulence (10, 11), and LOS also provides a second line of defense in ensuring its ability to survive in the host (12–14). The critical role of the Neisseria LOS in human pathogenesis has been demonstrated with the acapsular Neisseria gonococci, for which infection is initiated by molecular mimicry of host glycosphingolipid by the antigenically similar LOS on the extracellular surface of the bacteria (9, 15), effectively evading the killing cascade of the host immune response (16). This camouflage mechanism is dependent on the addition of a terminal sialic acid to the LOS that enhances binding of the host regulatory protein Factor H to the bacterial surface, conferring resistance to complement-mediated killing (2, 16).
Two major outer core oligosaccharide termini, globotriaose (Gal-α1,4-Gal-β1,4-Glc) and lacto-N-neotetraose (Gal-β1,4-GlcNAc-β1,3-Gal-β1,4-Glc), are sialylated in Neisseria LOS with identical counterparts found in the glycosphingolipids of cells and tissues mediating intercellular recognition in important biological processes in humans (9). The critical role of LOS sialylation in pathogenesis is also reflected by the constitutive expression and maintenance of a highly conserved LOS-specific sialyltransferase in the clinically invasive Neisseria pathogens (sequence identity >95%) (17, 18). The Neisseria LOS-specific sialyltransferase (NST) catalyzes the transfer of sialic acid (Neu5Ac) from the donor (CMP-Neu5Ac) to the non-reducing end of the galactosyl residue at the terminus of the LOS acceptor. In addition, NST is bifunctional in both acceptor specificity (e.g. the globotriaose- and lacto-N-neotetraose-containing LOS acceptors) and flexibility in catalyzing either α2,3 or α2,6 glycosidic bond formation (9). In N. meningitidis expressing the L1-type LOS and some Neisseria gonorrhoeae strains that express an acceptor LOS with a globotriaose terminus, NST catalyzes α2,6 glycosidic bond formation between the sialic acid and the terminal galactosyl residue on the acceptor (16, 19). In N. meningitidis serotypes L2, -3, -5, -7, and -9 and some N. gonorrhoeae strains, NST catalyzes α2,3 linkage formation from sialic acid to the lacto-N-neotetraose moiety on LOS (9).
Based on sequence relationships, sialyltransferases have been categorized into five different glycosyltransferase families (GT-29, -38, 42, -52, and -80) in the Carbohydrate Active enZYmes data base (CAZY) (20). Under this convention, NST is placed within the GT-52 family. The structures of the bacterial sialyltransferases are of biochemical and pharmaceutical interest because sialylation contributes directly to bacterial virulence, and the three-dimensional structure of sialyltransferases can provide insight into the design of specific inhibitors that have promise as potential antibacterials. The four previously reported sialyltransferase structures (from CAZY families GT29, -42, -80) have showed either of two types of protein fold typical of all nucleotide-activated glycosyltransferase families, denoted as GT-A and GT-B (21). The crystal structures of the LOS-specific sialyltransferases from Campylobacter jejuni (CST-I and -II) in family GT-42 is representative of the GT-A fold (22), whereas the two recently determined sialyltransferase structures from Pasteurella multocida (23–25) and Photobacterium phosphoreum (26) in family GT-80 employ a GT-B fold. In addition, NST has a predicted transmembrane helix at the N terminus that anchors the catalytic domain to the outer membrane on the extracellular surface (18). In this work we have developed a detergent-based purification strategy that significantly improves the solubility of an engineered form of NST from N. meningitidis, facilitating biochemical and crystallographic analysis. The NST structures and accompanying kinetic/mechanistic analysis of structure-based point mutations reveal many unique features that are distinct from previously published glycosyltransferases. In particular, observation of a domain-swapped dimeric form of the enzyme that appears to be mediated by lipid binding and the observed requirement for the presence of high affinity acceptor substrates and analogs for crystallization of the NST-donor complex could imply a potential activating role of the acceptor substrate in regulating NST function at the extracellular surface of the bacterial membrane.
EXPERIMENTAL PROCEDURES
The truncation mutant Δ29NST was constructed from the previously cloned N. meningitidis 126E(L1) LOS sialyltransferase structural gene (GenBankTM accession number U60662) (27). Two versions of the Δ29NST were constructed. One carries a thrombin-cleavable maltose-binding protein (MBP) fusion tag at the N terminus of Δ29NST in the pCW vector described previously (28). The other carries a thrombin-cleavable His8 tag at the C terminus of Δ29NST in the pET41b+ vector (Novagen), to which a thrombin-cleavable sequence was introduced in between the HindIII and NotI restriction sites. Site-directed mutagenesis was performed with the Pfu HT QuikChange kit (Stratagene) on both the MBP-fused Δ29NST structural gene in pCW vector and the His8-tagged Δ29NST structural gene in pET41 vector to make the following single point mutants: E124A (GAG → GCT), H280A (CAC → GCG), R282A (CGC → GCA), D258A (GAC → GCG), D258N (GAC → AAC), D164N (GAC → AAC), and D165N (GAC → AAC). The DNA sequences of the constructs and mutants were validated via DNA sequencing by the Nucleic Acid Protein Service Unit at the University of British Columbia (Vancouver, BC, Canada). All chemicals were purchased from Sigma. The substrate analogs, CMP-3F(axial)-Neu5Ac (a synthetic Neu5Ac with a fluorine atom substitution at axial) and CMP-3F(equatorial)-Neu5Ac were synthesized and purified using previously established methods (29).
Cloning, Expression, and Purification of the MBP-Δ29NST Fusion Protein
The Δ29NST structural gene was directly amplified by PCR from MBP-NST with the forward primer, 5′-GCTGGAGCTGGACATATGGGGGAAAGGAATGCGGTTTCCCTGC-3′, which contains a NdeI restriction site (underlined) in the 5′ end, and the reverse primer, 5′-CCTAGGTCGACTCATTAATTTTTATCGTCAAATGTCAAAATC-3′, which contains a SalI restriction site (underlined). The amplified PCR products were digested with NdeI and SalI and then ligated using T4 DNA ligase into the expression vector, pCWMalET, which encodes the MBP fusion domain for enhanced solubility and ease of purification. The plasmid product was transformed into Escherichia coli AD202 (E. coli Genetic Stock Center, CGSC #7297) using electroporation, and the Δ29NST was expressed in the form of the MBP fusion protein.
volume of the overnight starter culture was used to inoculate the 2x yeast extract-tryptone (2YT) media supplemented with 100 μg/ml ampicillin and 0.2% (w/v) of d(+)-glucose. The cultures were incubated with shaking (200 rpm at 37 °C) until an A600 of ∼ 0.2 and were induced by the addition of 1 mm isopropyl β-d-thiogalactopyranoside at 20 °C with further overnight incubation. Approximately 11 g of cell pellet was collected from 2 liters of overnight culture. Protein purification was carried out at 4 °C. The cell pellet was resuspended in lysis buffer (2 mm EDTA (pH 7.5), 20 mm Tris-HCl (pH 7.5), 200 mm NaCl, 10% (v/v) glycerol) supplemented with a Roche Applied Science EDTA-free protease inhibitor mixture tablet and lysed at 20,000 p.s.i. using the Avestin Emulsiflex homogenizer. The lysate was centrifuged at 27,000 × g for 30 min, and the resulting supernatant was loaded onto a column (1 column volume of 20 ml) of amylose resin (New England Biolabs) that was equilibrated with the binding buffer (2 mm EDTA (pH 7.5), 20 mm Tris-HCl (pH 7.5), 200 mm NaCl, 10% (v/v) glycerol, and 5 mm β-mercaptoethanol). The bound MBP-Δ29NST fusion protein was fractionated with the elution buffer (binding buffer plus 10 mm maltose). The MBP-Δ29NST was dialyzed overnight in 20 mm Tris-HCl (pH 8.0) and 10% (v/v) glycerol and then further purified on a Mono Q (GE Healthcare) ion exchange column equilibrated with the binding buffer (20 mm Tris-HCl (pH 8.0), and 10% (v/v) glycerol). Fractions with pure MBP-Δ29NST eluted between 150 and 250 mm NaCl. Last, the MBP-Δ29NST was concentrated on a Centricon 30-kDa cutoff membrane and buffer-exchanged to 20 mm Tris-HCl (pH 7.5), 200 mm NaCl, and 10% (v/v) glycerol over a Superdex 200 gel-filtration column (GE Health).
Cloning, Expression, and Purification of the Δ29NST
The primers for the Δ29NST destined for the pET-41b+ vector were 5′-GCATTACGCATATGGGGGAAAGGAATGC-3′ (NdeI restriction site is underlined) and 5′-GCATTACGAAGCTTATCGTCAAATGTCAAA-3′ (HindIII restriction site is underlined). The PCR reaction was performed using the MBP-Δ29NST in pCW vector as a template. The PCR products were restricted with NdeI and HindIII and then ligated to a modified pET-41b+ expression vector in which a thrombin-cleavable sequence has been introduced to facilitate removal of the His8 tag from expressed Δ29NST. The plasmid product was transformed into the expression strain E. coli BL21(DE3). volume of the overnight starter culture was used to inoculate the LB media. The culture was incubated with shaking at 200 rpm and 37 °C until the A600 reached 0.6 and then heat-shocked at 42 °C by air for 45 min (30). After heat shock, the culture was cooled to 20 °C in a water bath followed by the addition of isopropyl β-d-thiogalactopyranoside to 0.75 mm for induction (30) and was further incubated overnight with shaking at 200 rpm in a 20 °C incubator. Approximately 6 g of cell pellet was collected from a 1-liter culture. All protein purification steps were carried out at 4 °C. The cell pellet was resuspended in the lysis buffer (20 mm MES (pH 6.5), 500 mm NaCl, 0.090% (w/v) Triton X-100) supplemented with the Roche Applied Science EDTA-free protease inhibitor mixture tablet and lysed using a high pressure homogenizer (Avestin Emulsiflex). The lysate was clarified by centrifugation at 100,000 × g for 40 min. Expressed Δ29NST in the supernatant was bound to a Ni2+-charged Chelating Sepharose Fast Flow column (GE Healthcare). 10 column volumes of wash buffer (20 mm MES (pH 6.5), 50 mm imidazole, 500 mm NaCl, 0.0090% (w/v) Triton X-100) followed by 10 column volumes of re-equilibration buffer (20 mm MES (pH 6.5), 150 mm NaCl, and 0.0090% (w/v) Triton X-100) were applied to wash the protein-bound column. Δ29NST was fractionated with the elution buffer (20 mm MES (pH 6.5), 150 mm NaCl, 0.0090% (w/v) Triton X-100, and 50 mm EDTA (pH 8.0)). Digestion of the His8 tag was carried out with a final concentration of 0.5 mg/ml thrombin (Sigma) on the purified Δ29NST for 2 days at 4 °C (>80% digested). The thrombin-digested reaction was concentrated 35-fold on a Centricon 30-kDa cutoff centrifugal concentrator and then further purified on a Superdex 200 (HR 10/30, column volume of 24 ml, GE Healthcare) pre-equilibrated with 20 mm MES (pH 6.5), 150 mm NaCl, and 0.0045% (w/v) of Triton X-100. The eluted Δ29NST was concentrated 2-fold on a 30-kDa cutoff centrifugal concentrator maintaining the final Triton X-100 concentration at or below 0.009% (w/v) and then flash-frozen in liquid nitrogen for storage. The final concentration of Δ29NST is typically in the range of 3–5 mg/ml, which is suitable for crystallization. The selenomethionine-labeled Δ29NST was prepared using the culture media described by Doublié (31) and purified using the procedure as described above.
Crystallization of the Δ29NST
The crystallization and subsequent crystal manipulations were performed at 4 °C. Δ29NST was crystallized by vapor diffusion methods using 1-μl drops of protein as prepared above mixed with an equal volume of crystallization buffer from the well reservoir. Initial screens were performed with the selenomethionine labeled apo form of Δ29NST. Ordered crystals formed in 100 mm sodium acetate (pH 4.2–4.4) and 1.7 m di-ammonium sulfate (space group P21212; Table 1, column b). Crystallization of native (non-selenomethionine-containing) Δ29NST with CDP was also achieved in this same crystal form (Table 1, column c). Attempts to co-crystallize under the same conditions Δ29NST in complex with both the equatorial variant of the donor substrate analog CMP-3F(equatorial)-Neu5Ac (5 mm) and the acceptor, lactose (25 mm), resulted in density defining only CMP bound to NST (Table 1, column a). This suggests that the 3F(equatorial)-Neu5Ac of the donor was likely hydrolyzed and released during the crystallization process. Further co-crystallization of ternary complex structures was attempted with 25–50 mm various acceptors (see below) and 0.35 mm CMP-3F(axial)-Neu5Ac donor analog (which showed better resistance to hydrolytic cleavage than the equatorial counterpart in our kinetic studies). Despite the testing of a variety of acceptors including the higher affinity lactosides benzyl lactoside, thiophenyl lactoside, BODIPY lactoside, and n-propyl lactoside, no density corresponding to the acceptor was visualized in our resulting electron density maps. However, interestingly, the presence of these acceptors in the crystallization mix was necessary to generate the binary complex structures of intact donor CMP-3F(axial)-Neu5Ac bound to NST. Highly similar donor analog complexes crystallized in space group C2221 were generated in this way using either 50 mm n-propyl lactoside (Table 1, column d) or benzyl lactoside acceptors in 180 mm trisodium citrate, 5 mm MnCl2, and 15% (v/v) PEG4000 (pH 6.5) in the reservoir as well as with 25 mm thiophenyl lactoside acceptor in 180 mm trisodium citrate, 5 mm MnCl2, and 17% (v/v) PEG2000 (pH 6.5) in the reservoir.
TABLE 1.
Data collection and refinement statistics
Numbers in parentheses refer to the appropriate outer resolution shell. SeMet, selenomethonine; r.m.s.d., root mean square deviation from ideal geometry.
| Parameters | a | b | c | d |
|---|---|---|---|---|
| SeMet-NST·CMP complex | SeMet-NST Free enzyme | NST·CDP complex | NST·CMP-3F(axial)-Neu5Ac complex | |
| Ligand(s) used for co-crystallization | CMP-3F(equatorial)-Neu5Ac + Lactose | CDP | CMP-3F(axial)-Neu5Ac + n-propyl lactoside | |
| Ligand observed in the structure | CMP | CDP | CMP-3F(axial)-Neu5Ac | |
| Lipid binding site modeled witha | PEG | Triton X-100 | PEG | PEG |
| PDB entry | 2yk5 | 2yk4 | 2yk6 | 2yk7 |
| Data collection statistics | ||||
| Beamline | ALS 822 | ALS 822 | Rigaku MicroMax 007HF | Rigaku MicroMax 007HF |
| Wavelength (Å) | 0.980 | 0.980 | 1.54 | 1.54 |
| Resolution limits (Å) | 2.33 (2.41-2.33) | 1.94 (1.97-1.94) | 2.83 (2.88-2.83) | 2.18 (2.22-2.18) |
| Space group | P21212 | P21212 | P21212 | C2221 |
| Unit cell dimension (Å) | 85.7, 124.1, 41.9 | 86.6, 124.7, 41.9 | 86.1, 123.5, 41.7 | 42.8, 101.4, 166.6 |
| Unique reflections | 19,839 | 34,365 | 11,162 | 19,377 |
| Redundancy | 7.1 (7.1) | 8.8 (8.5) | 6.8 (5.1) | 7.6 (5.4) |
| Completeness (%) | 100.0 (100.0) | 100 (99.9) | 99.7 (97.8) | 97.1 (84.9) |
| I/σ(I) | 17.0 (3.89) | 35.5 (3.22) | 15.4 (2.70) | 23.9 (2.64) |
| Rmerge (%) | 13.7 (40.0) | 6.7 (46.2) | 12.1 (52.2) | 9.0 (46.7) |
| Mosaicity | 0.18-0.26 | 0.20-0.32 | 0.38-0.47 | 0.68-0.82 |
| Refinement statistics | ||||
| Resolution limits (Å) | 2.3 | 2.0 | 2.8 | 2.2 |
| Number of reflections in working set | 18,825 | 32,604 | 10,613 | 17,844 |
| Number of reflections in test set | 1,008 | 1,735 | 529 | 957 |
| Rfactor of working set (%) | 17.5 | 19.3 | 18.1 | 19.6 |
| Rfree (%) | 22.0 | 22.0 | 24.0 | 25.3 |
| r.m.s.d. bond length (Å) | 0.019 | 0.020 | 0.020 | 0.019 |
| r.m.s.d. bond angles (°) | 1.680 | 1.880 | 1.835 | 1.773 |
| Average B factors (Å2) | 20.7 | 33.0 | 28.4 | 30.4 |
| Ramachandran analysis (most favored, allowed, disallowed) | 98.2, 1.8, 0% | 99.4, 0.6, 0% | 95.0, 5.0, 0% | 97.8, 2.2, 0% |
a The head group of the detergent Triton X-100 is polyethylene glycol (PEG) composed of 9 or 10 units of ethylene glycol.
Structure Determination of Δ29NST
The initial model of the selenomethionine-incorporated Δ29NST (CMP-bound NST complex; Table 1, column a) was solved in the P21212 space group at 2.4 Å resolution from single wavelength anomalous dispersion data collected on the peak of the selenium absorption edge (0.9798 Å with an F′ of −8.0 and an F″ of 4.5) on beamline 822 at the Advanced Light Source (Berkeley, CA). Diffraction data were indexed and integrated with DENZO and reduced with SCALEPACK using the HKL2000 program suite (32). A calculated Matthews coefficient of VM = 2.79 Å3/dalton suggests 1 monomer of NST per asymmetric unit and a solvent content of 56%. The scaled data together with the amino acid sequence of the Δ29NST was input into the Phenix-AutoSol suite, which located 11 of the 12 selenium sites (Met-1 is not detected) and built 214 of the 340 residues (Rfactor of 40%). Improvement of the initial model was carried out interactively with the model building program Coot (33) and refined with the program Refmac in the CCP4 software suite (34). During refinement of the initial model, it became clearly evident from the observed electron density that a domain swap of the 2 monomers aligning on the 2-fold crystallographic symmetry axis was present as judged by the 2Fo − Fc composite omit map. The refined selenomethionine-labeled structure was subsequently used as a phasing model to solve the apo and binary complex structures of the ligand-bound NST in both space groups P21212 and C2221 using the program MolRep. The crystallographic asymmetric unit in the space groups C2221 and P21212 contains a monomer of NST in association with another subunit related by a crystallographic dyad axis of 180°, suggesting an extensive homo-dimerization of NST. This same homo-dimerization interface is also supported by lower resolution NST structures solved in space group P212121, for which the crystallographic asymmetric unit contains the two molecules of a similarly domain swapped dimer of NST (data not shown). We also note that in our electron density maps, residues Pro-49—Lys-370 and the engineered C-terminal thrombin cleavage sequence are clearly visible, but the segment from Gly-30 to Gln-48 is disordered, suggesting mobility of the N termini in the crystal lattice. In the four structures listed in Table 1, columns a–d, Ramachandran analysis (Procheck (35)) indicated that all the backbone ψ and φ angles are in the most favored (98.15, 99.38, 95.05, and 97.81%) and in the allowed (1.85, 0.62, 4.95, and 2.19%) conformations, respectively.
Activity Assays
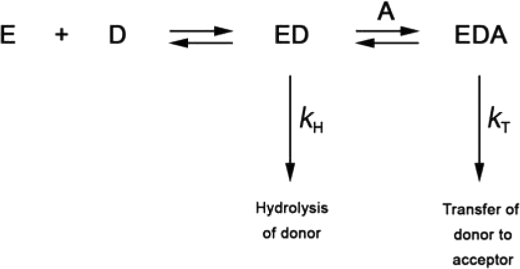
Sialyltransferase activity was monitored using a continuous coupled assay analogous to that previously described (22). However, without Triton X-100, Δ29NST (expressed and purified from the His8 tag fusion construct) is highly insoluble. Second, the solubility of Δ29NST is temperature-sensitive, and the protein precipitates at room temperature (23 °C). For the purpose of our activity and kinetic measurements, we therefore used a fusion of NST with MPB (MalE) MBP-Δ29NST, which remains soluble at the physiological temperature (37 °C) necessary for the coupling enzymes in the established assay procedure. Maintenance of folding and structural integrity of the purified MBP-Δ29NST and its single point mutants was confirmed by circular dichroism. Using our coupled enzyme assay, a significant level of spontaneous background hydrolysis of donor CMP-Neu5Ac by NST in the absence of acceptor substrate was observed. To ensure the accurate measurement of transferase activity, a stable rate of spontaneous hydrolysis of donor CMP-Neu5Ac was established before acceptor substrate was added to initiate assays. The competition for the NST-bound CMP-Neu5Ac in the enzyme-catalyzed hydrolysis reaction (kH) versus the formation of the ternary complex (NST·CMP-Neu5Ac· acceptor) in the transferase reaction (kT) gives rise to the reaction pathway illustrated in Scheme 1. Therefore, the hydrolase activity (kH) was subtracted from the observed enzyme activity (rate of CMP production) to obtain the transferase activity (kT). Kinetic parameters were determined by direct fit of the data to the relevant expression using the program GraFit.
SCHEME 1.
The reaction pathway of the NST-catalyzed reaction. Association of enzyme NST (E) and CMP-Neu5Ac donor substrate (D) forms the binary complex (ED). In the absence of acceptor substrate (A), NST spontaneously hydrolyzes the bound CMP-Neu5Ac at the rate constant, kH. In the presence of acceptor (A), the binary complex (ED) and acceptor substrate (A) form the ternary complex (EDA) and catalyze the transfer of Neu5Ac to the acceptor substrate at rate constant, kT.
RESULTS AND DISCUSSION
Overall Architecture of NST Mediates a Unique Domain-swapped Homodimerization
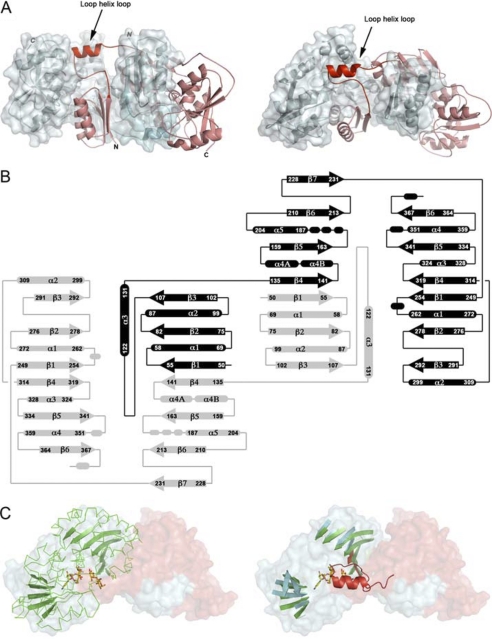
Wild type NST functions at the surface of the Neisseria outer membrane (18). Removal of the predicted transmembrane helix and the addition of the detergent Triton X-100 allowed us to create a solubilized and active form of the enzyme (Δ29NST) suitable for large-scale purification and formation of ordered crystals. The structure of Δ29NST was solved under different conditions in two distinct space groups to a maximum resolution of 1.94 Å (Table 1). The structures of Δ29NST in these space groups are highly similar in overall topology with a root mean square deviation of 0.1–3.5 Å2 (average of 1.3 Å2) on the 316 observed Cα atoms (calculated using CCP4-SUPERPOSE) and with the only significant differences due to localized conformational changes involved in binding the substrate analog in the binary complex captured in the C2221 crystal form. Importantly, in both space groups formation of a highly similar homodimer of NST is observed across the 2-fold symmetry axis (Fig. 1A).
FIGURE 1.
A, shown is the NST domain-swapped homodimer. The two polypeptide chains are colored in red (α) and blue (β). In addition, the β subunit is displayed with an opaque solvent-accessible surface to show the extensive contact surface for the α subunit. The N-terminal 29 amino acid residues corresponding to a transmembrane helix were truncated from the N terminus (labeled). The loop-helix-loop highlights the swapping hinge module (Leu-108—Lys-134). B, the secondary structure topology diagram is shown. The two polypeptide chains are shaded in black and gray. The labeled numbers correspond to the amino acid residues in the NST protein sequence. The secondary structures are labeled as α for α helices and β for β strands and are numbered successively in the polypeptide chain. Residues 108–134 correspond to the hinge loop module in the NST swapped dimer. C, PmST1 (P. multocida) sialyltransferase is superimposed to one-half of the NST dimer. Left, the domain-swapped NST homodimer is shown in a solvent-accessible surface representation and is colored in red for the peptide chain α and blue for the peptide chain β. PmST1 is shown in green ribbon with the central β-sheets highlighted as arrows. The co-crystallized CMP-3F(axial)-Neu5Ac and lactose in PmST1 are shown in yellow-red stick representation. Right, the green ribbons of PmST1 are omitted for clarity. The figure shows the superimposition of the central β sheets between the PmST1 and one functional subunit in NST. The swapping hinge loop module (loop-helix-loop) colored in red is superimposed to the acceptor lactose in the PmST1 complex structure.
The overall structure of each Δ29NST monomer contains two Rossmann nucleotide binding folds; thus the enzyme has a topology that resembles the GT-B-type glycosyltransferases (21). In Δ29NST, the N (Pro-49—Tyr-231) and C (Thr-249–Lys-370)-terminal regions are linked by a peptide loop (Leu-232—Gly-248) and separated by a deep crevice that contains the substrate binding site (Fig. 1). However, the structure is uniquely characterized by a prominent domain swap in this region involving the exchange of Pro-49—Met-130 between the two associating monomers. The N-terminal domain begins with a 3-stranded parallel β-sheet (Nβ1, Nβ2, and Nβ3), with helices Nα1 and Nα2 stacking on one side, and continues with an extensive loop structure (108–121) that projects the helix Nα3 away from the originating subunit core (Fig. 1B). Unlike the canonical GT-B fold in which the corresponding loop exiting the strand Nβ3 continues in the opposite direction and orients the helix Nα3 directly on the central β-sheet in the N-terminal domain, the helix Nα3 of NST emerges from the originating subunit and extends the following four-stranded parallel β-sheet structure (Nβ4, Nβ5, Nβ6, Nβ7) to dock alongside the three-stranded β sheet (Nβ1, Nβ2, and Nβ3) in the adjacent subunit, resulting in a composite parallel β-sheet with the strand order (3214567). This mixed central β-sheet is sandwiched by the helices Nα1 and Nα2 from the peptide chain on one side and the helices Nα4 and Nα5 of the second peptide chain on the other side, forming the swapped N-terminal domain. In addition, the exchanged subdomain (Pro-49—Met-130) also makes extensive molecular contacts with the C-terminal domain, which follows the conserved GT-B folding scheme involving formation of a seven-stranded central β-sheet with the strand order (321456) and which is sandwiched by the connecting helices of Cα1 and Cα2 on one side and helices Cα3 and Cα4 on the other side. The domain swap observed in our NST structures buries 3150 Å2 of molecular surface area at the dimer interface. 32 residues from each subunit contribute several dozen hydrogen bonds and 8 salt bridges across the dimer interface (calculated using PISA, European Bioinformatics Institute) (36). It has been suggested that domain swapping in protein structures allows for enhanced structural stability (37), a notion supported in NST by previous studies showing the maintenance of activity and dimerization in the presence of denaturing agents for detergent-solubilized NST isolated from bacterial membranes (38). In this work dimerization of Δ29NST in solution was further characterized by analytical gel-filtration and static light scattering (supplemental Fig. 2). In addition, chemical cross-linking with the homo-bi-functional amine cross-linkers (ethylene glycol bis[succinimidyl succinate] (EGS) and Dithiobis[succinimidyl propionate] (DSP) followed by MALDI-TOF mass spectrometry showed a molecular weight for the major peaks in the mass spectra of the EGS- and DSP-treated Δ29NST sample, consistent with dimer formation (80 kDa; supplemental Fig. 3). Similarly, the intact MBP-Δ29NST fusion utilized in our kinetic assays also forms a strong dimer in solution.
Lipid Binding Cavity in NST
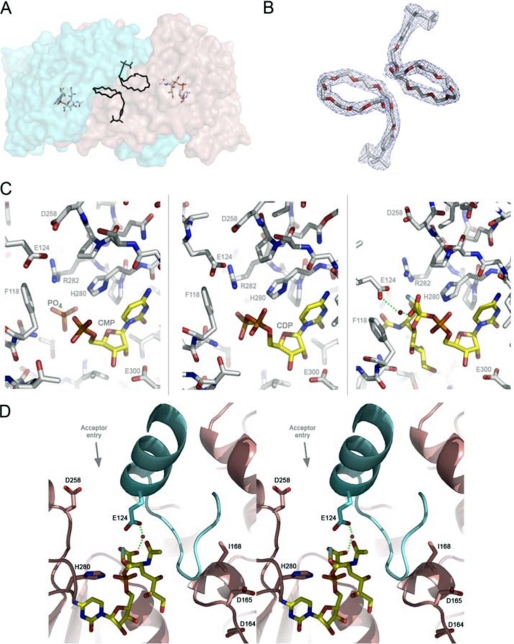
In the structure of Δ29NST, a tunnel-shaped hydrophobic cavity was observed between helix Nα4 and the loop (Leu-108—Thr-121) at the point of the domain swap. In all our structures, strong electron density corresponding to an extended (presumably) acyl chain runs along this hydrophobic tunnel. In the difference Fourier map (Fo − Fc) of Δ29NST, the extended electron density is tethered along the dimerization interface of the N-terminal domain such that the two extended molecules lie adjacent to each other across the 2-fold symmetry axis to stabilize the swapped dimeric form (Fig. 2, A and B). We predict these two linear chains belong to either the fatty acyl chains of a diglyceride lipid, captured in vivo from the bacterial membrane where NST is expressed, or the two polyethylene glycol groups of the detergent Triton X-100, which we have shown to be necessary for solubilization and activity of our overexpressed protein in vitro. NST is a membrane-bound enzyme that acts on a lipid-containing LOS acceptor. We propose that in vivo the diglyceride lipid would insert its fatty acyl chains (18–22 carbon-units-long) into each of the two functional subunits and stabilize the swapped dimerization necessary for NST activity and specificity.
FIGURE 2.
A, the lipid binding site modeled with the polyethylene glycol group of Triton X-100 in the NST homodimer is shown. The modeled polyethylene glycols are located at the dimerization interface at the approximate center of the NST dimer. For purpose of illustration, the Triton X-100 in the apoNST structure is superimposed into the NST·CMP-3F(axial)-Neu5Ac complex structure, and the distance and orientation to the bound CMP-3F(axial)-Neu5Ac are shown. B, the 2Fo − Fc electron density maps (contoured at 1 σ) fit with Triton X-100. C, the sialic acid binding site in NST is shown. The three structures of NST co-crystallized with structural donor sugar analogs or products (CMP, CDP, CMP-3F(axial)-Neu5Ac). In all three structures, the protein-ligand interactions are conserved (labeled). Phe-118 and Glu-124 reside on the swapped loop from the adjacent monomer. D) Stereo view of the catalytic machinery of NST. In contrast to the CAZY GT-80 sialyltransferases family in which the general base is part of the highly conserved (D/E)(D/E)G sequence motif, NST likely utilizes a distinct carboxylate, Asp-258, as a general base. The (D/E)(D/E)G motif (Asp-164—Asp-165—Gly-166 in NST) is separated from the active site by the swapped loop-helix-loop structure (colored in cyan) and appears to play a structural rather than catalytic role in the GT-52 family. Interestingly, point mutations at position 168, which is one residue away from the highly conserved (D/E)(D/E)G motif, has been shown to switch the transferase specificity between α2,3 and α2,6 linkage formation (27). Our structures of NST may explain this result in that such point mutations in or near the Asp-164—Asp-165—Gly-166 motif could have an indirect effect on NST function by altering the geometry of the adjacent swapped loop-helix-loop and the donor substrate binding residues therein (Glu-124 for example). This image was created in Wall-Eye Stereo mode using PyMOL.
Kinetic Analysis of NST
Using our previously described coupled enzyme assay (22), the NST kinetic properties obtained with the MBP-Δ29NST fusion protein (as described under “Experimental Procedures”) were measured under various conditions with a number of lactoside acceptors (Table 2). In the absence of acceptor substrate, NST hydrolyzes CMP-Neu5Ac; thus, hydrolysis rates were subtracted from overall turnover rates in our calculations as described under “Experimental Procedures.” Acceptor sugars containing aromatic aglycones are preferred substrates indicating the formation of additional binding interactions that mimic those ordinarily formed with sugars within the inner core-oligosaccharide region of LOS acceptor in the membrane.
TABLE 2.
Specificity of MBP-Δ29NST for aglycone-lactoside substrates
The error range in the data is 10–15%.
| Acceptor | Km | kcat | kcat/Km |
|---|---|---|---|
| mm | min−1 | mm−1min−1 | |
| Lactosea | 0.021 | ||
| LacNAc | 55 | 9 | 0.16 |
| Benzyl-β-lacNAc | 16 | 6 | 0.38 |
| Benzyl-β-lactoside | 39 | 3.6 | 0.09 |
| Phenyl-β-lactoside | 13 | 2.1 | 0.16 |
| Thiophenyl-β-lactoside | 7 | 2 | 0.28 |
| Propyl-β-lactosidea | 0.017 |
a Due to the high mm concentration for Km, only second order rates of the reaction were determined. Km values for acceptors were determined at a constant CMPNeuAc concentration of 0.4 mm.
In keeping with earlier work showing that Triton X-100 detergent supports activity in the membrane-associated NST (15, 38–40), our analysis shows that MBP-Δ29NST is active in the presence of Triton X-100 or CHAPS, whereas other detergents such as octyl glucoside, octyl glucoside, lauryldimethylamine-oxide (LDAO), Fos-choline-12, or n-dodecyl-N,N-dimethylglycine inactivates the enzyme completely. The maintenance of enzyme activity with Triton X-100 is compatible with association of Triton X-100 in the structure of Δ29NST. However, we also found that NST activity is dependent on Triton X-100 concentration; supplemental Fig. 1 shows a reactivation curve with a minimal activity near the Triton X-100 critical micelle concentration.
Mode of Donor CMP-3F(axial)-Sialic Acid Binding
In the binary complex structure of NST and the donor sugar analog CMP-3F(axial)-Neu5Ac, the CMP moiety is coordinated by conserved residues in the Rossmann-fold-containing C-terminal domain (Fig. 2C). The imidazole side chain of His-280 forms a hydrogen bond to the phosphate group, and the carboxylate side chain of Glu-300 forms a bidentate anchor to the C2 and C3 hydroxyl groups on the ribose ring. The cytidine ring is sandwiched between the alkyl side chains of Pro-281 and Ilw-299 and held by two hydrogen bonds from the backbone amide oxygen of Ala-278 and the backbone amide nitrogen of Ile-299 to the N4 nitrogen and C2 oxygen on the cytidine ring, respectively. Similarly, in the binary complex of CMP and NST, the product CMP anchors to the C-terminal domain of NST in the same manner as the CMP moiety of the donor analog in the binary complex of NST and CMP-3F(axial)-Neu5Ac (Fig. 2C).
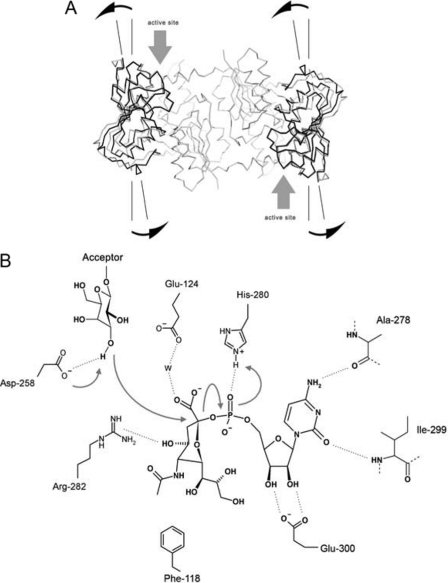
The sialic acid binding site of the donor sugar is located at the interface of the N and C domains and is positioned next to the swapped loop-helix-loop motif (Leu-108—Lys-134). The phenyl side chain of Phe-118 provides the platform on which the sialyl carboxyl group, through a bridging water molecule, forms a hydrogen bond to the side chain of Glu-124. The guanidinium group of Arg-282 is within hydrogen bonding distance of the C4 hydroxyl group on the sialic acid ring and is adjacent to the C3 fluorine substituent and the C5 N-acetyl moiety. It is interesting to note that both Glu-124 and Phe-118 originate from the adjacent subunit at the point of the domain swap, highlighting the necessarily composite nature of the active site in the NST homodimer (Fig. 2C), a feature characteristic common with other domain-swapped enzymes in which catalytic residues reside on separate peptide chains (41–46), and with additional ligand binding sites created at the dimerization interface (47–49). Together, the guanidinium group of Arg-282, the carboxyl side chain of Glu-124, the N-acetyl group of the donor sialic acid, the phenyl side chain of Phe-118, and the carboxyl side chain of Asp-258 form the periphery of the open entrance above the reaction center.
Catalytic Mechanism of NST
NST catalysis involves nucleophilic attack of the acceptor hydroxyl on the anomeric center of the donor substrate, with consequent inversion of stereochemistry at the C2 position of the donor sialic acid moiety. In our binary complex structure of donor CMP-3F(axial)-Neu5Ac and NST, the C2 anomeric center is exposed to a spacious cavity in the active site, which the acceptor sugar can access. Our structures show that the side chains of Glu-124 and Asp-258 are in close proximity to the C2 reaction center, both of which have the potential to act as a general base in activating the acceptor substrate nucleophile with the adjacent Arg-282 suitably positioned to play a role in binding of substrate and/or stabilization of reaction intermediates. As such, these residues were mutated to assess their functional roles in catalysis. The results in Table 3 support the observed interaction between Glu-124 and the sialic acid moiety in our binary complex structure as the E124A mutation weakens the binding affinity (but not catalytic rate) for the donor substrate. Mutation of Arg-282 to alanine (R282A), on the other hand, decreases both affinity and the rate of sialic acid transfer to the acceptor substrate, indicating not only the importance of the observed interaction with the sialic acid C4 OH in substrate-binding but as well a potential modulation of the electrostatic microenvironment by the guanidinium side chain that optimizes turnover. Mutation of Asp-258 to asparagine (D258N) results in an inactive mutant and completely deactivates both the hydrolytic pathway (kH) and transferase pathway (kT). In the structure of NST, Asp-258 is located in the loop between strand Cβ1 and helix Cα1 in the C-terminal domain and assumes two side-chain conformations in the structures solved, orienting the carboxylate side chain toward the bulk solvent space on the protein surface or toward the anomeric center of the 3F(axial)-Neu5Ac moiety (distance of ∼7 Å). Asp-258 could, therefore, potentially act as a molecular gate controlling access of the sialic acid moiety to the reaction center. Based on the spatial proximity and orientation of Asp-258 and the geometry of the bound CMP-3F(axial)-Neu5Ac in the binary complex structure, the complete loss of activity (kH and kT) upon mutation at this position suggests that Asp-258 would be potentially the strongest candidate for general base function in NST, albeit potentially requiring further (side chain and/or acceptor induced) conformational changes to bring it into the <3 Å range appropriate to support such a role. Clearly future structures with acceptor bound will be needed to validate the nature of the general base directly. It should be noted that although completely conserved in the Neisseria subgroup of the GT-52 family, sequence variability in this localized region in other uncharacterized or non-sialyltransferase members of GT-52 would suggest an alternate base may be utilized in the differing functions and catalytic mechanisms of these variants (for example the GT-52 retaining α-glucosyltransferase, WaaH (50)).
TABLE 3.
Catalytic efficiencies and substrate binding constants of MBP-Δ29NST and single point mutants in the hydrolysis and sialyltransfer reactions
| Hydrolysis of CMP-Neu5Ac in the absence of acceptor substrates | |||||
| Kinetic parameters | MBP-29NST | R282A | E124A | H280A | D258Na |
| kcat (min−1) | 0.59 | 0.32 | 0.55 | 0.05 | – |
| Km (μm) | 20 | 55 | 29 | 12.7 | – |
| kcat/Km (mm−1min−1) | 29 | 5.76 | 18.7 | 3.9 | – |
| Transfer of CMP-Neu5Ac to acceptor substrates | |||||
| Kinetic parameters | MBP-29NST | R282A | E124A | H280A | D258Na |
| kcat (min−1) | 5.4 | 0.62 | 4.4 | 0.108 | – |
| Km (μm) | 43 | 62 | 194 | 54 | – |
| kcat/Km (mm−1min−1) | 124 | 10 | 23 | 2 | – |
a –, D258N mutant had no activity in either enzyme-catalyzed hydrolysis of CMPNeuAc or enzyme-catalyzed transfer of CMPNeuAc.
Mutation of His-280 (H280A), whose side chain interaction with the phosphate group of CMP-Neu5Ac is conserved in the P. multocida GT-80 sialyltransferase (24), decreases rates of both hydrolysis of CMP-Neu5Ac (kH) and of transfer (kT) (Scheme 1; Table 3). This significant decrease in catalytic efficiency is compatible with the general acid function proposed for the corresponding histidine residue (His-311) in the P. multocida sialyltransferase (24), and these two histidine residues (His-280 in NST, and His-311 in PmST1) superimpose closely in the structural alignment. In contrast to the general acid function, the general base function in NST is significantly different from that of PmST1. In PmST1, the general base function resides on the highly conserved (D/E)(D/E)G sequence motif (24, 26, 51); however, this conserved signature sequence (Asp-164—Asp-165—Gly-166 in NST) is involved in stabilizing the fold of the N-terminal domain of NST and is not localized within the active site of NST (Fig. 2D) (i.e. these consensus motifs do not align with each other in either space or in structure). We have constructed the corresponding D164N and D165N mutants of this motif in the MBP-Δ29NST for kinetic characterization and in the His8-tagged Δ29NST for crystallization. However, both D164N and D165N mutants are inactive with respect to hydrolysis (kH) and transferase activities (kT). We suggest that the complete loss of function is likely the result of changes in the structural integrity of the enzyme as the D164N formed insoluble inclusion bodies upon expression and the D165N single point mutants failed to form crystals. At the same time, the inactive mutant of our postulated general base, D258N, exhibited similar solubility properties to native protein and in our high resolution analysis of the mutant by x-ray crystallography was found to have the same domain-swapped dimer structure. Our analysis of active site single point mutants reveals that both the general base (Asp-258) and general acid (His-280) are located on the C-terminal domain of NST and that the enzyme utilizes a conventional nucleophilic displacement mechanism, likely via a somewhat “exploded” oxocarbenium ion-like transition state as shown in Fig. 3B.
FIGURE 3.
A, conformational change upon CMP-3F(axial)-Neu5Ac binding is shown. Comparison of the apoNST and the donor analog-bound structures reveals an approximate 10° rotation on a linear axis formed by the Cαs of Leu-253, Val-277, Tyr-318, and Tyr-337 localized to the central β-sheet in the C-terminal domain using the model-building program Coot. This conformational change effectively pries open the N- and C- interdomain cleft (highlighted with gray arrows), making the active site more accessible to substrates. B, shown is the proposed catalytic mechanism for NST. The transfer reaction occurs between the terminal galactosyl residue of the acceptor substrate and the anchored sialic acid moiety that is covalently linked to the rigidly coordinated CMP group in NST. This CMP binding mode is strengthened by bifurcated hydrogen bonds between Glu-300 and the ribosyl hydroxyl groups and by the specific recognition of the cytidine base that is established through hydrogen bonds with the main chain amide and carbonyl groups of Ile-299 and Ala-278, respectively. The unique geometry of the sialic acid moiety in the CMP-3F(axial)-Neu5Ac·NST complex is stabilized through several interactions including a hydrogen bond from the His-280 side chain to the CMP phosphate of the donor, a water-mediated hydrogen bond bridging the sialyl carboxylate to the side chain of Glu-124, and a hydrogen bond between the C4 hydroxyl group of sialic acid and the guanidinyl group of Arg-282. These interactions fix the apparent half-chair sialyl ring geometry in NST and orient the anomeric center toward the entrance of the active site cavity, ready for transfer to the acceptor substrate. Activation of the acceptor nucleophile (deprotonation of the hydroxyl group on the galactosyl acceptor) is carried out by the proposed general base Asp-258, which is located at the rim of the active site entrance and next to the C2 anomeric center on sialic acid. A concerted mechanism is presented in the figure and follows the classical SN2 type nucleophilic substitution reaction, which involves the development of sp2-like hybridization at the sialyl C2 position, and formation of a planar oxocarbenium ion in the transition state. Inversion of stereochemistry at the C2 reaction center that leads to formation of an α-configured glycosidic bond in the sialylated galactosyl product is enhanced by general acid assistance from the proximal His-280, which stabilizes the developing negative charge on the phosphate leaving group.
Potential Implications for Acceptor Binding
In terms of acceptor sugar binding, the helix Nα3 and the flanking loops (Leu-108—Lys-134) that mediate the domain swap in the NST homo-dimer are likely, based on the evidence below, to constitute the major interaction site (Fig. 1A). In this site the loop Leu-108—Thr-121 (localized between strand Nβ3 and helix Nα3) is unusually extended, allowing it to be sandwiched between the C termini of the central β-sheet and the helix Nα3 and with packing interactions along helix Nα5 in the N-terminal domain (Fig. 1B). Compellingly, comparison with other monomeric GT-B glycosyltransferases in the CAZY data base shows that the equivalent loop structure is much smaller (∼7–10 residues shorter) and cannot form the equivalent packing interactions to those seen in our domain-swapped NST. Interestingly, however, these shortened loops have been shown directly by crystallographic analysis to be involved in binding acceptor substrates in the monomeric GTs (52–57) including two recently determined sialyltransferases in the GT-80 family, PmST1 from P. multocida and Psp26ST from P. phosphoreum (24, 26). In superpositions of the domain-swapped NST with these latter monomeric complex structures, the bound lactose acceptor is positioned immediately next to the Leu-108—Lys-134 loop of NST, suggesting that this point of domain swapping is a unique structural feature that functions in specifically binding the lipidated acceptor substrates of these Neisseria GT52 sialyltransferases (Fig. 1C).
Furthermore, our data may suggest that the complex multicomponent nature of native LOS acceptors could well have multiple roles in NST function: the lipid A tails binding our observed lipid binding channels to mediate NST dimerization and formation of the active site; the inner core oligosaccharides playing a further “activation role” to pry open the unusually closed NST active site for subsequent donor binding; finally, the binding of the more terminal outer core oligosaccharides as the direct acceptor for glycosyltransfer. The proposed activation role could be a form of regulatory control at the bacterial membrane and would be compatible with a catalytic mechanism involving the ordered addition of substrates as outlined in Scheme 1. The concept is supported by co-crystallization experiments of the donor-NST complex structures in which we observed that only the presence of aglycone/hydrophobic lactoside acceptor variants in the crystallization mix allowed for the formation of ternary complexes with sialic acid containing donor analogs (all other more simple acceptors tried including lactose, galactose, and N-acetyl galactosamine failed to produce such complexes). Furthermore, in our NST structures, the N and C domains are tightly associated in the free enzyme form, with the active site occluded from bulk solvent; it is only upon the addition of the aglycone/hydrophobic lactoside acceptors in the crystallization mix that the interdomain space is expanded to accommodate the donor sugar, CMP-3F(axial)-Neu5Ac (Fig. 3A) in the complex structure, suggesting a region of these complex acceptors apart from the direct point of transfer is responsible for the requisite conformational opening to allow donor binding. This represents a unique conformational requirement among the currently known GT-B-fold glycosyltransferases, which typically adopt an open conformation in the free enzyme form and a closed compact state in the substrate-bound structures (21, 58). It is intriguing to correlate these novel features/observations, as they imply an activation function played by the lipidated acceptor substrates in the structure and function of NST. Because of the increased affinity and specificity (kcat/Km) for aglycone/hydrophobic substitutions on the reducing terminus of acceptor lactosides (Table 2), the interaction, and specifically, the contribution/recognition of aglycone substitutions on the reducing terminus of lactosides to enzyme activation highlights the potential role of the equivalent inner/outer core carbohydrate clusters of LOS in the activation of NST function. Together with the presence of the occupied lipid-like binding cavities that we observe adjacent to the predicted NST acceptor site, our data provide significant new insight into how the GT-52 family of sialyltransferases has uniquely evolved to promote interactions with the lipidated LOS acceptor substrate in the bacterial membrane.
Acknowledgments
We thank Dr. Michel Gilbert and Lisa Willis for contributions to earlier stages of this project. We thank the X-ray Crystallography Hub at the Centre for Blood Research (University of British Columbia) and the Advanced Light Source (Lawrence Berkeley National Laboratory, Berkeley, CA) for data collection.
This work was supported by Canadian Institutes of Health Research Operating Grant MOP84272 (to N. C. J. S., W. W. W., and S. G. W.), the Howard Hughes Medical Institute International Scholar Program (to N. C. J. S.), scholarship funding from the Michael Smith Foundation for Health Research (to L. Y.-C. L. and C. P. C. C.), postdoctoral fellowship from the NSERC (to B. R.), and infrastructure funding from the Canada Foundation for Innovation and the British Columbia Knowledge Development Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
The atomic coordinates and structure factors (codes 2yk5, 2yk4, 2yk6, 2yk7) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- LOS
- lipooligosaccharide
- MBP
- maltose-binding protein
- GT
- glycosyltransferase
- NST
- N. meningitidis serotype L1.
REFERENCES
- 1. Angata T., Varki A. (2002) Chem. Rev. 102, 439–469 [DOI] [PubMed] [Google Scholar]
- 2. Schneider M. C., Prosser B. E., Caesar J. J., Kugelberg E., Li S., Zhang Q., Quoraishi S., Lovett J. E., Deane J. E., Sim R. B., Roversi P., Johnson S., Tang C. M., Lea S. M. (2009) Nature 458, 890–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ram S., Sharma A. K., Simpson S. D., Gulati S., McQuillen D. P., Pangburn M. K., Rice P. A. (1998) J. Exp. Med. 187, 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lasky L. A. (1995) Annu. Rev. Biochem. 64, 113–139 [DOI] [PubMed] [Google Scholar]
- 5. Cebo C., Dambrouck T., Maes E., Laden C., Strecker G., Michalski J. C., Zanetta J. P. (2001) J. Biol. Chem. 276, 5685–5691 [DOI] [PubMed] [Google Scholar]
- 6. Troy F. A., 2nd (1992) Glycobiology 2, 5–23 [DOI] [PubMed] [Google Scholar]
- 7. Yuki N. (2000) Cell. Mol. Life Sci. 57, 527–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Houliston R. S., Koga M., Li J., Jarrell H. C., Richards J. C., Vitiazeva V., Schweda E. K., Yuki N., Gilbert M. (2007) Biochemistry 46, 8164–8171 [DOI] [PubMed] [Google Scholar]
- 9. Moran A. P., Prendergast M. M., Appelmelk B. J. (1996) FEMS Immunol. Med. Microbiol. 16, 105–115 [DOI] [PubMed] [Google Scholar]
- 10. Mandrell R. E., Apicella M. A. (1993) Immunobiology 187, 382–402 [DOI] [PubMed] [Google Scholar]
- 11. Swartley J. S., Marfin A. A., Edupuganti S., Liu L. J., Cieslak P., Perkins B., Wenger J. D., Stephens D. S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 271–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jennings M. P., Hood D. W., Peak I. R., Virji M., Moxon E. R. (1995) Mol. Microbiol. 18, 729–740 [DOI] [PubMed] [Google Scholar]
- 13. Preston A., Mandrell R. E., Gibson B. W., Apicella M. A. (1996) Crit. Rev. Microbiol. 22, 139–180 [DOI] [PubMed] [Google Scholar]
- 14. Kahler C. M., Martin L. E., Shih G. C., Rahman M. M., Carlson R. W., Stephens D. S. (1998) Infect. Immun. 66, 5939–5947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mandrell R. E., Smith H., Jarvis G. A., Griffiss J. M., Cole J. A. (1993) Microb. Pathog. 14, 307–313 [DOI] [PubMed] [Google Scholar]
- 16. Gulati S., Cox A., Lewis L. A., Michael F. S., Li J., Boden R., Ram S., Rice P. A. (2005) Infect. Immun. 73, 7390–7397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rest R. F., Mandrell R. E. (1995) Microb. Pathog. 19, 379–390 [DOI] [PubMed] [Google Scholar]
- 18. Shell D. M., Chiles L., Judd R. C., Seal S., Rest R. F. (2002) Infect. Immun. 70, 3744–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wakarchuk W. W., Gilbert M., Martin A., Wu Y., Brisson J. R., Thibault P., Richards J. C. (1998) Eur. J. Biochem. 254, 626–633 [DOI] [PubMed] [Google Scholar]
- 20. Coutinho P. M., Deleury E., Davies G. J., Henrissat B. (2003) J. Mol. Biol. 328, 307–317 [DOI] [PubMed] [Google Scholar]
- 21. Lairson L. L., Henrissat B., Davies G. J., Withers S. G. (2008) Annu. Rev. Biochem. 77, 521–555 [DOI] [PubMed] [Google Scholar]
- 22. Chiu C. P., Watts A. G., Lairson L. L., Gilbert M., Lim D., Wakarchuk W. W., Withers S. G., Strynadka N. C. J. (2004) Nat. Struct. Mol. Biol. 11, 163–170 [DOI] [PubMed] [Google Scholar]
- 23. Ni L., Sun M., Yu H., Chokhawala H., Chen X., Fisher A. J. (2006) Biochemistry 45, 2139–2148 [DOI] [PubMed] [Google Scholar]
- 24. Ni L., Chokhawala H. A., Cao H., Henning R., Ng L., Huang S., Yu H., Chen X., Fisher A. J. (2007) Biochemistry 46, 6288–6298 [DOI] [PubMed] [Google Scholar]
- 25. Kim D. U., Yoo J. H., Lee Y. J., Kim K. S., Cho H. S. (2008) BMB Rep. 41, 48–54 [DOI] [PubMed] [Google Scholar]
- 26. Kakuta Y., Okino N., Kajiwara H., Ichikawa M., Takakura Y., Ito M., Yamamoto T. (2008) Glycobiology 18, 66–73 [DOI] [PubMed] [Google Scholar]
- 27. Wakarchuk W. W., Watson D., St. Michael F., Li J., Wu Y., Brisson J. R., Young N. M., Gilbert M. (2001) J. Biol. Chem. 276, 12785–12790 [DOI] [PubMed] [Google Scholar]
- 28. Wakarchuk W. W., Campbell R. L., Sung W. L., Davoodi J., Yaguchi M. (1994) Protein Sci. 3, 467–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu H., Yu H., Karpel R., Chen X. (2004) Bioorg. Med. Chem. 12, 6427–6435 [DOI] [PubMed] [Google Scholar]
- 30. Chen J., Acton T. B., Basu S. K., Montelione G. T., Inouye M. (2002) J. Mol. Microbiol. Biotechnol. 4, 519–524 [PubMed] [Google Scholar]
- 31. Doublié S. (1997) Methods Enzymol. 276, 523–530 [PubMed] [Google Scholar]
- 32. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 33. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 35. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 36. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 37. Qu C., Liljas L., Opalka N., Brugidou C., Yeager M., Beachy R. N., Fauquet C. M., Johnson J. E., Lin T. W. (2000) Structure 8, 1095–1103 [DOI] [PubMed] [Google Scholar]
- 38. Gilbert M., Cunningham A. M., Watson D. C., Martin A., Richards J. C., Wakarchuk W. W. (1997) Eur. J. Biochem. 249, 187–194 [DOI] [PubMed] [Google Scholar]
- 39. Izumi M., Shen G. J., Wacowich-Sgarbi S., Nakatani T., Plettenburg O., Wong C. H. (2001) J. Am. Chem. Soc. 123, 10909–10918 [DOI] [PubMed] [Google Scholar]
- 40. Gilbert M., Watson D. C., Cunningham A. M., Jennings M. P., Young N. M., Wakarchuk W. W. (1996) J. Biol. Chem. 271, 28271–28276 [DOI] [PubMed] [Google Scholar]
- 41. Sun W., Singh S., Zhang R., Turnbull J. L., Christendat D. (2006) J. Biol. Chem. 281, 12919–12928 [DOI] [PubMed] [Google Scholar]
- 42. Hare S., Bayliss R., Baron C., Waksman G. (2006) J. Mol. Biol. 360, 56–66 [DOI] [PubMed] [Google Scholar]
- 43. Gunawan J., Simard D., Gilbert M., Lovering A. L., Wakarchuk W. W., Tanner M. E., Strynadka N. C. J. (2005) J. Biol. Chem. 280, 3555–3563 [DOI] [PubMed] [Google Scholar]
- 44. Liaw S. H., Chang Y. J., Lai C. T., Chang H. C., Chang G. G. (2004) J. Biol. Chem. 279, 35479–35485 [DOI] [PubMed] [Google Scholar]
- 45. Hadden J. M., Convery M. A., Déclais A. C., Lilley D. M., Phillips S. E. V. (2001) Nat. Struct. Biol. 8, 62–67 [DOI] [PubMed] [Google Scholar]
- 46. Chen Y., Narendra U., Iype L. E., Cox M. M., Rice P. A. (2000) Mol. Cell 6, 885–897 [PubMed] [Google Scholar]
- 47. Fromme R., Katiliene Z., Giomarelli B., Bogani F., Mc Mahon J., Mori T., Fromme P., Ghirlanda G. (2007) Biochemistry 46, 9199–9207 [DOI] [PubMed] [Google Scholar]
- 48. Calarese D. A., Scanlan C. N., Zwick M. B., Deechongkit S., Mimura Y., Kunert R., Zhu P., Wormald M. R., Stanfield R. L., Roux K. H., Kelly J. W., Rudd P. M., Dwek R. A., Katinger H., Burton D. R., Wilson I. A. (2003) Science 300, 2065–2071 [DOI] [PubMed] [Google Scholar]
- 49. Lewis R. J., Muchová K., Brannigan J. A., Barák I., Leonard G., Wilkinson A. J. (2000) J. Mol. Biol. 297, 757–770 [DOI] [PubMed] [Google Scholar]
- 50. Kaniuk N. A., Monteiro M. A., Parker C. T., Whitfield C. (2002) Mol. Microbiol. 46, 1305–1318 [DOI] [PubMed] [Google Scholar]
- 51. Freiberger F., Claus H., Günzel A., Oltmann-Norden I., Vionnet J., Mühlenhoff M., Vogel U., Vann W. F., Gerardy-Schahn R., Stummeyer K. (2007) Mol. Microbiol. 65, 1258–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mulichak A. M., Losey H. C., Walsh C. T., Garavito R. M. (2001) Structure 9, 547–557 [DOI] [PubMed] [Google Scholar]
- 53. Mulichak A. M., Losey H. C., Lu W., Wawrzak Z., Walsh C. T., Garavito R. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9238–9243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mulichak A. M., Lu W., Losey H. C., Walsh C. T., Garavito R. M. (2004) Biochemistry 43, 5170–5180 [DOI] [PubMed] [Google Scholar]
- 55. Bolam D. N., Roberts S., Proctor M. R., Turkenburg J. P., Dodson E. J., Martinez-Fleites C., Yang M., Davis B. G., Davies G. J., Gilbert H. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5336–5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mittler M., Bechthold A., Schulz G. E. (2007) J. Mol. Biol. 372, 67–76 [DOI] [PubMed] [Google Scholar]
- 57. Hu Y.., Chen L., Ha S., Gross B., Falcone B., Walker D., Mokhtarzadeh M., Walker S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 845–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Newcomer M. E., Lewis B. A., Quiocho F. A. (1981) J. Biol. Chem. 256, 13218–13222 [PubMed] [Google Scholar]