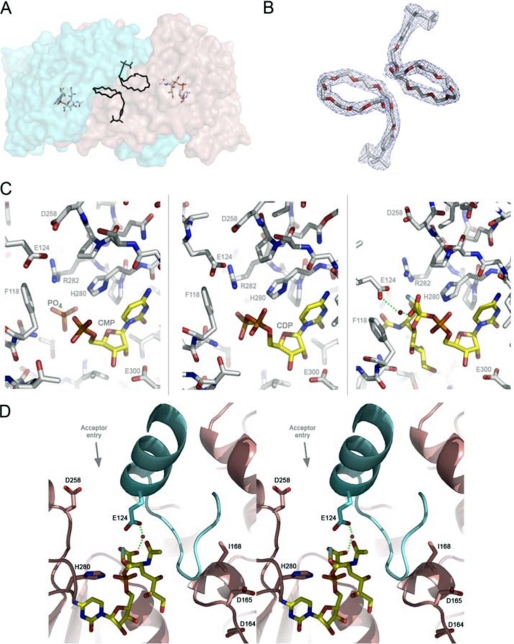
FIGURE 2.
A, the lipid binding site modeled with the polyethylene glycol group of Triton X-100 in the NST homodimer is shown. The modeled polyethylene glycols are located at the dimerization interface at the approximate center of the NST dimer. For purpose of illustration, the Triton X-100 in the apoNST structure is superimposed into the NST·CMP-3F(axial)-Neu5Ac complex structure, and the distance and orientation to the bound CMP-3F(axial)-Neu5Ac are shown. B, the 2Fo − Fc electron density maps (contoured at 1 σ) fit with Triton X-100. C, the sialic acid binding site in NST is shown. The three structures of NST co-crystallized with structural donor sugar analogs or products (CMP, CDP, CMP-3F(axial)-Neu5Ac). In all three structures, the protein-ligand interactions are conserved (labeled). Phe-118 and Glu-124 reside on the swapped loop from the adjacent monomer. D) Stereo view of the catalytic machinery of NST. In contrast to the CAZY GT-80 sialyltransferases family in which the general base is part of the highly conserved (D/E)(D/E)G sequence motif, NST likely utilizes a distinct carboxylate, Asp-258, as a general base. The (D/E)(D/E)G motif (Asp-164—Asp-165—Gly-166 in NST) is separated from the active site by the swapped loop-helix-loop structure (colored in cyan) and appears to play a structural rather than catalytic role in the GT-52 family. Interestingly, point mutations at position 168, which is one residue away from the highly conserved (D/E)(D/E)G motif, has been shown to switch the transferase specificity between α2,3 and α2,6 linkage formation (27). Our structures of NST may explain this result in that such point mutations in or near the Asp-164—Asp-165—Gly-166 motif could have an indirect effect on NST function by altering the geometry of the adjacent swapped loop-helix-loop and the donor substrate binding residues therein (Glu-124 for example). This image was created in Wall-Eye Stereo mode using PyMOL.