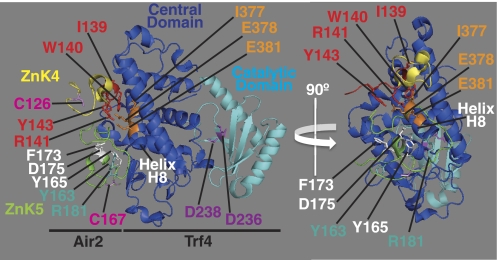
FIGURE 8.
Trf4-Air2 structural model that highlights Air2 ZnK4, ZnK5, and IWRXY motif residues and Trf4 central and catalytic domain residues. A structural model of the Trf4-Air2 complex (Protein Data Bank code 3NYB) based on the recent work of by Hamill et al. (49) is shown to highlight positions of key amino acid residues analyzed in this study. Two views of the Trf4-Air2 complex rotated 90° about the vertical axis relative to each other are shown. Air2 ZnK4 (yellow), Air2 ZnK5 (green), Trf4 central domain (blue), and Trf4 catalytic domain (cyan) are depicted. Air2 ZnK4 residue Cys-126 and ZnK5 Cys-167 (pink), equivalent to Air1 Cys-139 and Cys-178, are labeled. Air2 ZnK5 residues Tyr-165, Phe-173, and Asp-175 (white), equivalent to Air1 ZnK5 residues Tyr-176, Phe-184, and Asp-186, and Air2 ZnK5 residues Tyr-163 and Arg-181 (teal), equivalent to Air1 ZnK5 residues Phe-174 and Arg-192, are labeled. Air2 IWRXY motif residues Ile-139, Trp-140, Arg-141, and Tyr-143 (red), equivalent to Air1 IWRXY residues Ile-152, Trp-153, Arg-154, and Tyr-156, are labeled. Trf4 central domain helix H8 (white) and helix H8 residues Ile-377, Glu-378, and Glu-381 (orange) are labeled. Trf4 catalytic residues Asp-236 and Asp-238 (magenta) are labeled. The structural model of the crystal structure of Trf4-Air2 fragment complex was reproduced from the Protein Data Bank file (entry 3NYB) of the atomic coordinates and structure factors (49) using MacPyMOL software (64) and altered and annotated using Adobe Photoshop and Illustrator CS4 (Adobe).