Abstract
Estrogen sulfotransferase (SULT1E1) catalyzes the sulfonation of estrogens, which limits estrogen mitogenicity. We recently reported that SULT1E1 expression is low in preconfluent MCF10A human breast epithelial cells but increases when the cells become confluent. Pulse-chase labeling experiments with 5-bromouridine demonstrated that the confluence-mediated increase in SULT1E1 expression was due to increased mRNA synthesis. Because aryl hydrocarbon receptor (AhR) activation has been shown to suppress SULT1E1 expression and loss of cell-cell contact has been shown to activate the AhR in other cell types, we tested whether the confluence-associated changes in SULT1E1 expression were mediated by the AhR. Relative to confluent MCF10A cells, preconfluent cells had higher levels of CYP1A1 mRNA and greater activation of an AhR-responsive luciferase reporter, demonstrating that the AhR was active in the preconfluent cells. AhR and aryl hydrocarbon receptor nuclear translocator mRNA and protein levels were also higher in preconfluent than in confluent cultures. Treatment of preconfluent cells with the AhR antagonist, 3′-methoxy-4′-nitroflavone (MNF), or AhR knockdown significantly increased SULT1E1 expression. MCF10A cells stably transfected with a luciferase reporter containing ∼7 kilobases of the SULT1E1 5′-flanking region showed both MNF- and confluence-inducible luciferase expression. Preconfluent cells transiently transfected with the reporter showed both MNF treatment- and AhR knockdown-mediated luciferase induction, but mutation of a computationally predicted dioxin response element (DRE) at nucleotide (nt) −3476 did not attenuate these effects. These results demonstrate that SULT1E1 expression in MCF10A cells is transcriptionally regulated by confluence through a suppressive action of the AhR, which is not mediated through a DRE at nt −3476.
Introduction
The SULTs are a family of conjugating enzymes that catalyze the transfer of a sulfuryl moiety from the activated physiological sulfate donor 3′-phosphoadenosine-5′-phosphosulfate to the hydroxyl groups of endogenous and xenobiotic substrates, including hormones, drugs, and procarcinogens (Falany, 1997; Glatt, 2000). One of the SULTs, SULT1E1, catalyzes the sulfonation of estrogens at physiological concentrations. SULT1E1 is an important determinant of a cell's response to estrogen because sulfonated estrogens cannot bind to ERs (Qian et al., 2001). In this manner, SULT1E1 expression in breast epithelial cells probably limits the mitogenic effects of estrogen, thereby reducing the risk for breast cancer development (Falany et al., 1995). SULT1E1 is expressed in human breast epithelial cells as well as in the MCF10A cell line, a model of normal human breast epithelial cells, but is down-regulated in many breast cancer cell lines, suggesting that this brake against estrogen mitogenicity is often lost during neoplastic transformation (Falany and Falany, 1996; Fu et al., 2010).
We recently reported that SULT1E1 mRNA content is markedly increased when replicating MCF10A cells become confluent (Fu et al., 2010), indicating that SULT1E1 expression is regulated according to the confluence of these cells. In comparison, in an earlier study in which we profiled the expression of cytochrome P450 transcripts in MCF10A cells, two cytochromes P450, CYP1A1 and CYP1S1, were expressed in preconfluent MCF10A cells but not in confluent MCF10A cells (Thomas et al., 2006). Because both of these cytochromes P450 are transcriptional targets of the AhR (Rivera et al., 2002), this finding suggests that the AhR is active in preconfluent MCF10A cells but inactive in confluent MCF10A cells.
We and others have previously reported that AhR agonist treatments cause suppression of SULTs in hepatic systems. In this regard, treatment of female rats with 3-methylcholanthrene caused suppression of hepatic hydroxysteroid sulfotransferase expression in parallel with CYP1A1 induction (Runge-Morris and Wilusz, 1994), and treatment with β-naphthoflavone or TCDD caused suppression of hydroxysteroid sulfotransferase and aryl sulfotransferase expression in primary cultured rat hepatocytes (Runge-Morris, 1998). In a microarray analysis of TCDD treatment effects on global gene expression in HepG2 human hepatoma cells, SULT1E1 mRNA content was decreased by 60% after treatment with 10 nM TCDD for 8 h (Puga et al., 2000). Approximately the same magnitude of suppression occurred when the cells were pretreated with cycloheximide before TCDD treatment, indicating that the reduction of SULT1E1 mRNA content was a direct effect of TCDD treatment on gene transcription and was not due to induction of a suppressive factor (Puga et al., 2000). Furthermore, TCDD treatment was reported to cause suppression of SULT1E1 expression in the livers of female C57BL/6 mice (Alnouti and Klaassen, 2008).
Taken together, these prior findings prompted us to hypothesize that the AhR is the molecular switch that confers confluence-dependent expression of SULT1E1 in MCF10A cells. We propose that basally active AhR suppresses SULT1E1 transcription in preconfluent MCF10A cells, whereas in confluent cells the AhR becomes inactive, thereby derepressing SULT1E1 transcription. In this study, we characterize the confluence dependence of SULT1E1 expression and investigate the role of the AhR in regulating this phenomenon.
Materials and Methods
Materials.
TCDD was purchased from Midwest Research Institute (Kansas City, MO). MNF was purchased from ICC Chemical Corporation (New York, NY). Cell culture medium, horse serum, l-glutamine, penicillin-streptomycin solution, sodium pyruvate, Lipofectamine 2000, TRIzol reagent, recombinant human SULT1E1, and anti-SULT1E1 antibody were purchased from Invitrogen (Carlsbad, CA). Epidermal growth factor was purchased from BD Biosciences (San Jose, CA). Recombinant human insulin (Novolin R) was purchased from Novo Nordisk Pharmaceuticals, Inc. (Princeton, NJ). BrU, cholera toxin, doxycycline, hydrocortisone, and puromycin were purchased from Sigma-Aldrich (St. Louis, MO). Mouse monoclonal AhR antibody (B-11), goat polyclonal ARNT1 antibody (C-19), rabbit polyclonal GAPDH antibody (FL-335), and horseradish peroxidase-conjugated goat anti-rabbit IgG, goat anti-mouse IgG, and donkey anti-goat IgG were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). ECL Plus Western Blotting Detection Reagents and Hybond-P membranes were purchased from GE Healthcare (Chalfont St. Giles, Buckinghamshire, UK). Other materials were obtained from the sources indicated below.
Cell Culture.
The MCF10A cell line was obtained from the Cell Resources Facility of the Barbara Ann Karmanos Cancer Institute, Wayne State University (Detroit, MI) and cultured in phenol red-free Dulbecco's modified Eagle's medium/Ham's F12 Nutrient Mixture (1:1) supplemented with 10 μg/ml insulin, 20 ng/ml epidermal growth factor, 100 ng/ml cholera toxin, 0.5 μg/ml hydrocortisone, 5% horse serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. The cell line was routinely maintained in T75 flasks in a 37°C humidified environment of 5% CO2-95% air. For a typical experiment, 125,000 cells or 1,000,000 cells were plated into 60-mm dishes (cell numbers for different vessel formats were adjusted according to their surface areas). At these cell densities, approximately 3 days after plating, ∼70% confluence (defined as preconfluence) or confluence was reached, respectively, and the cells were harvested for preparation of total RNA. For TCDD or MNF treatment, 48 h after plating, cells were treated with 0.1% DMSO (control), TCDD, or MNF for 24 h.
Gene Expression Analysis.
Total RNA was prepared from individual dishes of cells using an RNeasy Mini Kit (QIAGEN, Valencia, CA). RNA samples (1.5 μg) were reverse-transcribed using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Transcript levels were measured using the following TaqMan Gene Expression Assays: Hs00193690_m1 (SULT1E1), Hs00169233_m1 (AhR), Hs00231048_m1 (ARNT), and Hs00153120_m1 (CYP1A1) (Applied Biosystems). Each PCR included 2 μl of reverse transcription reaction as template, a primer/probe (5-carboxyfluorescein fluor-minor groove binder quencher) set, a primer-limited primer/probe (VIC fluor-minor groove binder quencher) set for 18S rRNA, and Universal PCR Master Mix, and amplifications were performed using a StepOnePlus Real-Time PCR System (Applied Biosystems). Thermocycling parameters were 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. Cycle threshold (Ct) values were obtained using the SDS software package (Applied Biosystems). For each sample, ΔCt was obtained by subtracting the Ct of 18S rRNA from the Ct of target mRNA. Then, ΔΔCt values were calculated by subtracting the ΔCt of the calibrator to which the other samples were compared from the ΔCt of each sample. Mean relative expression values were then calculated as 2−ΔΔCt.
For measuring the amount of SULT1E1 hnRNA, total RNA was isolated using an RNeasy Mini Kit with on-column DNase I treatment, and samples of total RNA (1.5 μg) were reverse-transcribed using a High Capacity cDNA Reverse Transcription Kit. As negative controls, equivalent amounts of total RNA were “mock reversed transcribed” by performing the reactions in the absence of reverse transcriptase. PCR primers were designed using Oligo Primer Analysis Software (version 7.36; Molecular Biology Insights, Cascade, CO) and the human SULT1E1 structural gene sequence (National Center for Biotechnology Information Reference Sequence NC_000004, 70706930–70725870 complement). The sequence of the upper primer (5′-GCTGGTCATCCAAATCCTG-3′) was located within exon 5 and the sequence of the lower primer (5′-CAATTTGCCTTCTACATCTGGACA-3′) was located within intron 5. Each PCR contained 1 μl of reverse transcription reaction as template, 25 μl of 2× SYBR Green PCR Master Mix (Applied Biosystems), and a 300 nM concentration each of upper and lower primer in a volume of 50 μl. Samples were incubated at 95°C for 10 min, followed by 35 cycles of 95°C for 15 s and 60°C for 1 min, followed by a melting curve of 95°C for 15 s, 60°C for 1 min, ramp to 95°C with data collection every 0.3°C, and 95°C for 15 s to ensure that a single product had been amplified. A commercial SYBR Green-based RT-PCR assay to detect TATA box binding protein was used for normalization (QIAGEN). After data acquisition, Ct values were determined, and data were analyzed as described above. Control reactions containing aliquots of the mock reverse-transcribed samples were performed to determine whether any fluorescent signal was derived from contaminating genomic DNA. To confirm amplification of the specific target fragment of expected size (203 nt), PCR products were run on a 2% agarose gel and visualized with ethidium bromide under ultraviolet illumination.
BrU Pulse-Chase Labeling.
MCF10A cells grown to preconfluence or confluence were incubated with 2 mM BrU in conditioned medium for 30 min to label nascent RNA. Cells were then washed 3 times in PBS and either collected directly (0-h time point) or chased in conditioned medium containing 20 mM uridine for 2 or 6 h at 37°C. Total RNA was isolated using TRIzol reagent, and the BrU-containing RNA was isolated using magnetic beads (Dynabeads, goat anti-mouse IgG; Invitrogen) conjugated to anti-BrdU monoclonal antibody (BD Biosciences). Conversion of the isolated BrU-containing mRNA into cDNA and real-time PCR analyses were performed by the Microarray Core of the University of Michigan Comprehensive Cancer Center (Ann Arbor, MI), according to protocols supplied by the manufacturer (SABiosciences, Frederick, MD). For the real-time PCR analyses, the Cancer Drug Resistance and Metabolism Real-Time RT PCR array (SABiosciences) and the ABI 7900HT Sequence Detection System from Applied Biosystems were used. The data were analyzed using RT2 Profiler PCR Array Data Analysis software (http://www.sabiosciences.com/pcr/arrayanalysis.php), and the data were normalized to the expression of five housekeeping genes present on the arrays. The housekeeping genes were β2-microglobulin (B2M), hypoxanthine phosphoribosyltransferase 1 (HPRT1), ribosomal protein L13A (RPL13A), GAPDH, and β-actin (ACTB).
Western Blot Analysis.
For measurement of SULT1E1 protein content, MCF10A cells in T75 flasks were washed with and scraped into ice-cold PBS. Cells were pelleted and homogenized by sonication in buffer (200 μl/flask) consisting of 50 mM Tris-HCl, 25 mM sucrose, 1 mM EDTA, and 1× Halt protease inhibitor (Thermo Fisher Scientific, Waltham, MA), pH 7.4. Homogenates were centrifuged at 20,000g and 4°C for 20 min, and supernatants were used for Western blot analysis. Protein concentrations were measured using the BCA Protein Assay (Thermo Fisher Scientific). Western blot analysis was performed as described previously (Fu et al., 2010), using 60 μg of sample protein, SULT1E1 antibody at a dilution of 1:2000, secondary antibody at a dilution of 1:10,000, and enhanced chemiluminescence for immunoreactive protein detection. For Western blot analysis of AhR and ARNT, MCF10A cells in 10-cm dishes were washed twice with PBS and then lysed using 700 μl of cold radioimmunoprecipitation assay buffer (Thermo Fisher Scientific). For SDS-PAGE, 30-μg samples of the lysates were separated on 4 to 20% Precise Protein Gels (Thermo Fisher Scientific). After transfer to polyvinylidene difluoride membranes, the blots were developed using AhR antibody (1:500) or ARNT antibody (1:400) and secondary antibodies at 1:10,000 dilutions. Western blots were normalized for variations in protein loading and transfer by redevelopment with a GAPDH antibody (1:1000).
MCF10A Cells Stably Expressing an AhR-Responsive Reporter.
MCF10A cells were plated into six-well plates (700,000 cells/well). The following day, 4 μg of pGudLuc1.1 (Garrison et al., 1996) (provided by Dr. Michael Denison, University of California, Davis, CA), which contains 4 DREs, and 0.65 μg of pSV2neo (American Type Culture Collection, Manassas, VA) were cotransfected into MCF10A cells using Lipofectamine 2000. After a 24-h recovery in standard medium, transfected cells were replated into medium containing 550 μg/ml G418. After two rounds of limiting dilution cloning, individual cell clones were identified and expanded. For TCDD treatment, 50,000 or 500,000 cells were plated into six-well plates, and 48 h after plating cells were treated with 0.1% DMSO or TCDD for 24 h. After treatment, growth medium was removed, and cells were washed with PBS. Cells were harvested by adding Passive Lysis Buffer (Promega, Madison, WI) to the wells (500 μl/well), and firefly luciferase activities were measured in lysate samples corresponding to 10 μg of cellular protein, estimated by harvesting replicate wells of cells in radioimmunoprecipitation assay buffer and measuring protein concentrations with the BCA Protein Assay. Firefly luciferase activities were measured using the Dual Luciferase Reporter Assay System (Promega) and an LMAX II384 microplate reader (Molecular Devices Corporation, Sunnyvale, CA) equipped with SoftMax Pro software.
Conditional Knockdown of AhR in MCF10A Cells.
A plasmid expressing a microRNA-adapted short hairpin RNA targeting human AhR in a doxycycline-inducible manner (oligo ID V2THS_132482, vector pTRIPZ) was purchased from Thermo Fisher Scientific, Open Biosystems Products (Huntsville, AL). For transfection, 700,000 MCF10A cells were plated into six-well plates, and 4 μg/well plasmid was transfected into the cells using Lipofectamine 2000 24 h after plating. Stably transfected cells were obtained by incubation in culture medium containing 1 μg/ml puromycin followed by limiting dilution cloning. To achieve AhR knockdown, cells were plated at low confluence and treated with 1 μg/ml doxycycline for 96 h, after which they were approximately 70% confluent.
Transfection of a SULT1E1 5′-Flanking Region-Luciferase Reporter Plasmid.
A fragment of the human SULT1E1 gene spanning from nt −7073 to +13 was amplified by PCR using genomic DNA from the MCF10A cell line as template [forward primer 5′-GGGGGTACCATTTGGCCTGCTATAACTGTATGCT-3′ (underlined sequence is a KpnI site) and reverse primer 5′-GGGCTCGAGACTTCTGCATTTGGAATGTTTCTGG-3′ (underlined sequence is a XhoI site)]. The amplified fragment was ligated into the KpnI and XhoI sites of the pGL4.17[luc2/Neo] reporter plasmid (Promega). The sequence of the SULT1E1 fragment was verified using the services of the Applied Genomics Technology Center, Wayne State University.
For stable transfection, 700,000 MCF10A cells were plated into six-well plates. The following day, the cells were transfected with 4 μg of the SULT1E1 5′-flanking region-luciferase reporter plasmid using Lipofectamine 2000. After a 24-h recovery in standard medium, the transfected cells were replated into medium containing 550 μg/ml G418 and expanded.
For transient transfections, MCF10A cells or MCF10A cells engineered for conditional knockdown of AhR were treated with either 0.1% DMSO or 1 μg/ml doxycycline for 96 h and were then subcultured into 24-well plates (150,000 cells/well). For transfection, the standard medium was replaced with serum-free Opti-MEM containing a premixed complex of 4 μl of Lipofectamine 2000, 0.8 μg of SULT1E1-luciferase reporter, and 1.25 ng of pRL-SV40 (Promega). The next day, the cultures (three wells per treatment group) were washed three times with fresh standard medium to remove dead cells and were then incubated with fresh medium containing 0.1% DMSO, 1 to 3 μM MNF, or 1 μg/ml doxycycline for 24 h. Under these transfection and treatment conditions, the cells were preconfluent throughout the experiment. The cells were then harvested for measurement of firefly and Renilla luciferase activities using the Dual Luciferase Reporter Assay System.
Computational Analysis of SULT1E1 5′-Flanking Region for DREs and Site-Directed Mutagenesis.
The region of the human SULT1E1 gene spanning from 10 kb upstream of the transcription start site through exon 1 was retrieved from the National Center for Biotechnology Information (nt 70,725,767 through 70,735,870 of NC_000004) and was evaluated for the presence of DREs using MatInspector (Genomatix Software, Ann Arbor, MI) (Cartharius et al., 2005). The V$AHRR (AHR-arnt heterodimers and AhR-related factors) matrix family was used for the search, and sites were considered to be matches if the calculated matrix similarity was greater than the optimized matrix threshold.
A single nucleotide change (C to A) was introduced into the core region of a DRE predicted to be located at nt −3476 of the SULT1E1 gene within the context of the luciferase reporter plasmid containing 7073 nt of the SULT1E1 5′-flanking sequence. This nucleotide change has been shown to abolish the ability of the AhR · ARNT heterodimer to bind to a DRE (Cuthill et al., 1991). The nucleotide change was introduced using the QuickChange II XL Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA), with forward primer 5′-ACAGCAAAAACCTGGGAGTGCATGTGCACACAC-3′ and reverse primer 5′-GTGTGTGCACATGCACTCCCAGGTTTTTGCTGT-3′ (the italicized sequence is the predicted DRE, the bolded sequence is the core region of the DRE, and the underscored nucleotide is the site of the mutation). The presence of the mutation was confirmed by sequence analysis.
Statistical Analysis.
Data were analyzed using the paired t test or one-way analysis of variance followed by the Newman-Keuls test using Prism (GraphPad Software Inc., San Diego, CA).
Results
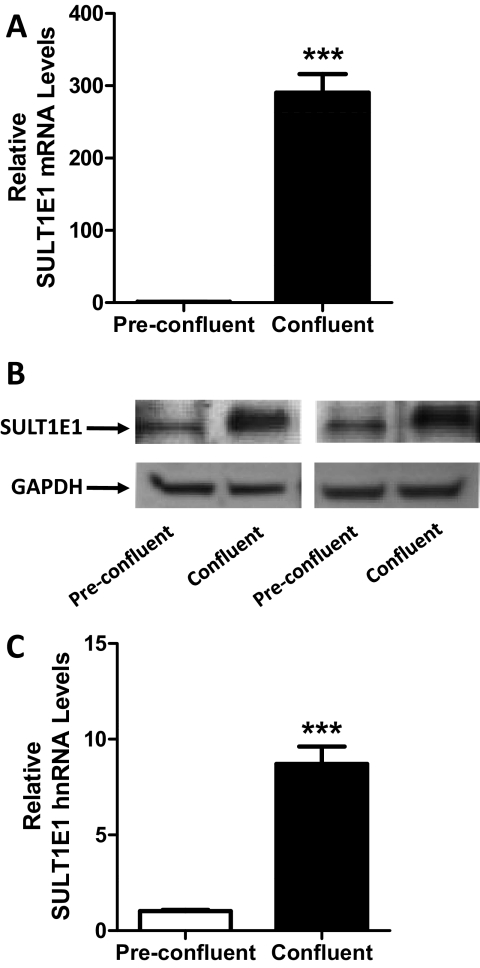
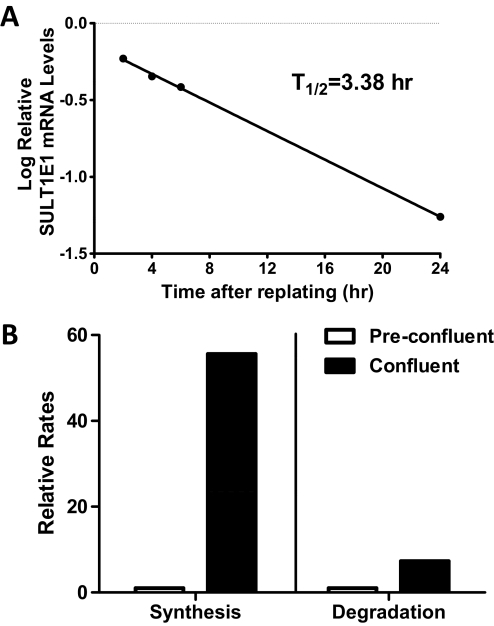
We previously reported that SULT1E1 mRNA was expressed at a higher level in confluent MCF10A cells than in preconfluent cells (Fu et al., 2010). To characterize the phenomenon more fully, we first confirmed the initial finding and then demonstrated that confluence-mediated up-regulation of SULT1E1 expression occurred at the protein level (Fig. 1, A and B). We next addressed whether the confluence-mediated increase in SULT1E1 mRNA content was the result of increased mRNA synthesis or decreased mRNA degradation. The relative levels of SULT1E1 hnRNA were measured in preconfluent and confluent MCF10A cells as an approximation of transcription rate. SULT1E1 hnRNA levels were significantly higher (∼8.7-fold) in confluent than in preconfluent MCF10A cells (Fig. 1C). By measuring SULT1E1 mRNA content at different times after replating of confluent MCF10A cells, the half-life of SULT1E1 mRNA in preconfluent cells was estimated to be 3.4 h (Fig. 2A). We used a newly described bromouridine labeling technique to measure the relative rates of SULT1E1 mRNA synthesis and degradation in preconfluent and confluent MCF10A cells (M. Paulsen and M. Ljungman, manuscript submitted for publication). By this analysis, the rate of SULT1E1 mRNA synthesis was 55.6-fold higher in confluent than in preconfluent cells, whereas the rate of SULT1E1 mRNA degradation was 7.3-fold higher in confluent than in preconfluent cells (Fig. 2B). Taken together, these results indicate that increased SULT1E1 mRNA stability cannot account for the increased SULT1E1 mRNA content that is seen upon cell confluence and that confluence-mediated up-regulation is most likely due to increased transcription.
Fig. 1.
SULT1E1 expression in preconfluent and confluent MCF10A cells. MCF10A cells were harvested at approximately 70 and 100% confluence. A, SULT1E1 mRNA levels were measured in six independent experiments using a TaqMan Gene Expression Assay. B, SULT1E1 immunoreactive protein levels were measured in two independent experiments by Western blot hybridization. C, SULT1E1 hnRNA levels were measured in six independent experiments using a SYBR Green Real-Time RT-PCR assay. In A and C, data are expressed as means ± S.E.M. ***, p < 0.001 compared with preconfluent cells.
Fig. 2.
Relative rates of SULT1E1 mRNA synthesis and degradation in preconfluent and confluent MCF10A cells. A, confluent MCF10A cells were subcultured and harvested at the indicated times after plating for measurement of SULT1E1 mRNA levels. SULT1E1 mRNA content is expressed as the log of the fractional level measured in confluent MCF10A cells, and the first-order half-life was calculated from the least-squares line. B, BrU labeling was used to measure relative rates of SULT1E1 mRNA synthesis and degradation in preconfluent and confluent MCF10A cells. The data represent averages from two independent experiments.
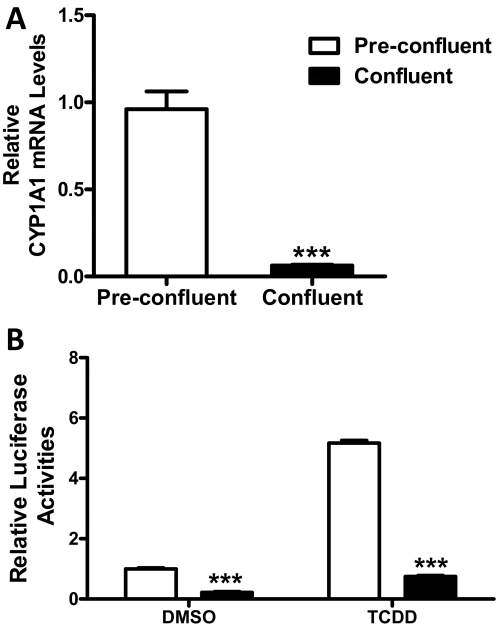
As described in the Introduction, confluence-mediated SULT1E1 up-regulation is mirrored by down-regulation of two cytochromes P450, CYP1A1 and CYP1S1, that are known transcriptional targets of the AhR (Thomas et al., 2006), suggesting that the AhR might provide the mechanistic link between these phenomena. Using real-time RT-PCR, we confirmed that CYP1A1 mRNA content was significantly higher in preconfluent MCF10A cells than in confluent cells (Fig. 3A). To evaluate further whether AhR activity varies as a function of MCF10A confluence, the cells were engineered to express firefly luciferase under the control of four DREs. TCDD treatment significantly increased luciferase reporter expression (5.2- and 3.4-fold in preconfluent and confluent cells, respectively), demonstrating responsiveness of the engineered cells to AhR activation (Fig. 3B). Luciferase expression was significantly (∼4.5-fold) higher in preconfluent cells than in confluent cells, supporting the conclusion that the AhR is basally active in preconfluent MCF10A cells but less active in confluent cells (Fig. 3B).
Fig. 3.
Indices of AhR activity in preconfluent and confluent MCF10A cells. A, MCF10A cells were harvested at approximately 70 and 100% confluence, and CYP1A1 mRNA levels were measured using a TaqMan Gene Expression Assay. Data are expressed as the mean ± S.E.M. of three independent experiments. B, MCF10A cells stably expressing an AhR-responsive luciferase reporter (pGudLuc1.1) were treated for 24 h with 0.1% DMSO or 30 nM TCDD before harvest at approximately 70 and 100% confluence for measurement of firefly luciferase activity and protein content. Data are expressed as mean relative luciferase activities (normalized to cellular protein content) ± S.D., three wells per treatment group. ***, p < 0.001 compared with preconfluent cells.
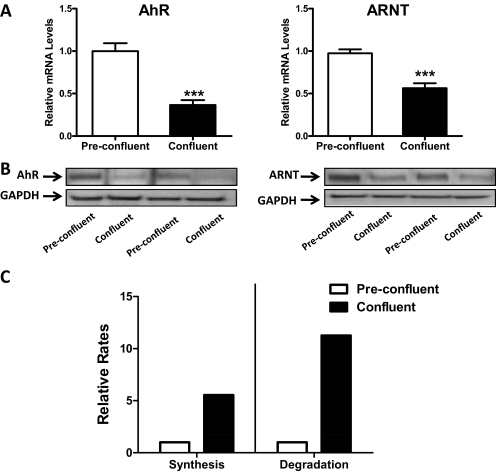
The mRNA levels of AhR and its heterodimerization partner ARNT were ∼3.0- and ∼1.8-fold higher, respectively, in preconfluent than in confluent cells, and the immunoreactive protein levels were correspondingly higher in preconfluent cells (Fig. 4, A and B). The higher levels of AhR mRNA and protein in preconfluent than confluent cells are probably attributable to differences in mRNA stability, because, using the aforementioned bromouridine labeling technique, the rates of AhR mRNA synthesis and degradation were determined to be 5.5- and 11.3-fold higher, respectively, in confluent MCF10A cells than in preconfluent cells (Fig. 4C).
Fig. 4.
Expression of AhR and ARNT in preconfluent and confluent MCF10A cells. A, MCF10A cells were harvested at approximately 70 and 100% confluence. AhR and ARNT mRNA levels were measured in six independent experiments using TaqMan Gene Expression Assays. Data are expressed as means ± S.E.M. ***, p < 0.001 compared with preconfluent cells. B, AhR and ARNT immunoreactive protein levels were measured in two independent experiments by Western blot hybridization. C, BrU labeling was used to measure relative rates of AhR mRNA synthesis and degradation in preconfluent and confluent MCF10A cells. The data represent the averages from two independent experiments.
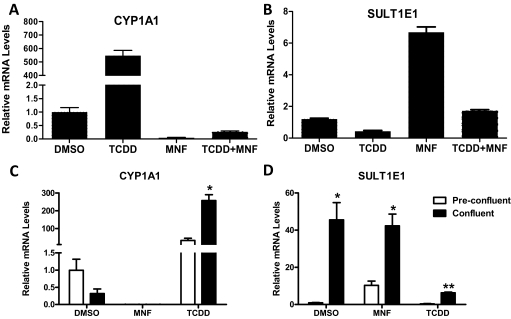
To investigate the role of the AhR in regulating SULT1E1 expression, we tested the effects of TCDD, a potent AhR agonist, and MNF, an AhR antagonist, on CYP1A1 and SULT1E1 expression in MCF10A cells. In preconfluent cells, 30 nM TCDD treatment increased CYP1A1 mRNA content by 547-fold, whereas treatment with 1 μM MNF, a concentration sufficient to abolish TCDD-mediated induction, decreased basal CYP1A1 mRNA content by >99% (Fig. 5A). In comparison, TCDD treatment of preconfluent MCF10A cells decreased SULT1E1 mRNA content by ∼57%, whereas MNF treatment increased the amount of SULT1E1 mRNA by ∼6.7-fold (Fig. 5B).
Fig. 5.
Effects of AhR agonist and antagonist treatments on CYP1A1 and SULT1E1 expression in MCF10A cells. A and B, preconfluent cultures of MCF10A cells were treated for 24 h with 0.1% DMSO, 30 nM TCDD, 1 μM MNF, or TCDD and MNF in combination and harvested for measurement of CYP1A1 (A) and SULT1E1 (B) mRNA levels. Data are expressed as the mean ± S.E.M. of triplicate assays. C and D, preconfluent and confluent cultures of MCF10A cells were treated for 24 h with DMSO, MNF, or TCDD and harvested for measurement of CYP1A1 (C) and SULT1E1 (D) mRNA levels. Data are expressed as the mean ± S.E.M. of three independent cell culture experiments. **, p < 0.01; *, p < 0.05 compared with preconfluent cells.
In a comparison of the effects of TCDD and MNF treatments in preconfluent MCF10A cells with those in confluent cells, TCDD-mediated CYP1A1 mRNA induction in confluent cells was not attenuated relative to the induction seen in preconfluent cells, despite the lower levels of AhR and ARNT, demonstrating the high efficacy of this AhR agonist. As shown before, basal levels of CYP1A1 mRNA were ∼69% lower in confluent cells than in preconfluent cells, although this difference was not significant when analyzed across three independent experiments (Fig. 5C). MNF treatment abolished basal CYP1A1 expression in both preconfluent and confluent cells (Fig. 5C). For SULT1E1, mRNA levels were significantly higher in confluent cells, and TCDD treatment suppressed expression in both preconfluent and confluent cells, again mirroring the effects seen for CYP1A1. MNF treatment increased SULT1E1 expression in preconfluent cells but not in confluent cells (Fig. 5D), suggesting that confluence and MNF treatment increase SULT1E1 expression through a common mechanism.
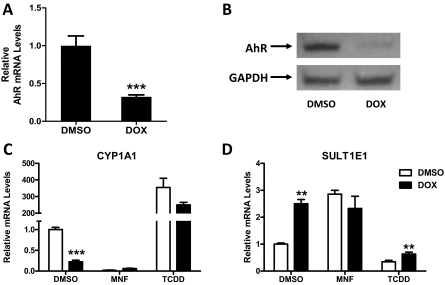
As a complementary approach, MCF10A cells were engineered for conditional (doxycycline-mediated) knockdown of AhR expression. A significant reduction of AhR mRNA content (by ∼68%) (Fig. 6A) and a marked decrease in the AhR immunoreactive protein level (Fig. 6B) were achieved when the engineered cells were treated with doxycycline for 96 h. This reduction in the AhR protein level was accompanied by only a ∼30% reduction in TCDD-inducible CYP1A1 expression (Fig. 6C) (not significant when analyzed across three independent experiments), again demonstrating the high efficacy of TCDD. When preconfluent cells were treated for 96 h with doxycycline, CYP1A1 mRNA levels were significantly reduced (by ∼78%), whereas SULT1E1 mRNA levels were significantly increased (by ∼2.5-fold) (Fig. 6, C and D). The doxycycline-mediated increase in SULT1E1 mRNA content was comparable to the increase that was produced when cells with intact AhR expression (i.e., not treated with doxycycline) were treated with MNF. In addition, cotreatment with doxycycline and MNF did not produce an additive effect, indicating that the effects of doxycycline and MNF on SULT1E1 expression were mediated through the common mechanism of AhR disruption.
Fig. 6.
Effect of AhR knockdown on CYP1A1 and SULT1E1 expression in preconfluent MCF10A cells. A and B, MCF10A cells engineered for conditional knockdown of AhR were plated at low density, treated for 96 h with 0.1% DMSO or 1 μg/ml doxycycline (DOX), and harvested at ∼70% confluence for measurement of AhR mRNA (A) and immunoreactive protein (B) levels. In A, data are expressed relative to DMSO-treated cells as the mean ± S.E.M. of three independent cell culture experiments. ***, p < 0.001 compared with DMSO-treated cells. C and D, MCF10A cells plated at low density were treated for 96 h with DMSO or doxycycline; during the final 24 h of this treatment period, the cells were additionally treated with DMSO, 1 μM MNF, or 30 nM TCDD. The cells were then harvested at ∼70% confluence for measurement of CYP1A1 (C) and SULT1E1 (D) mRNA levels, and data are expressed as the mean ± S.E.M. of three independent cell culture experiments. ***, p < 0.001; **, p < 0.01 compared with DMSO-treated cells.
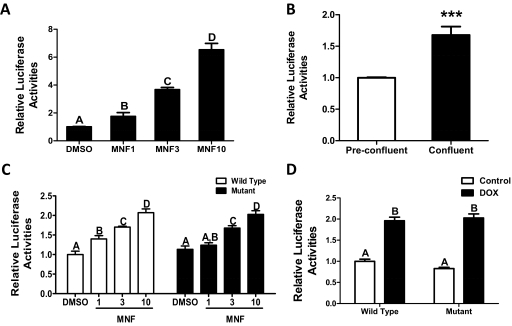
Computational analysis of 10 kb of the SULT1E1 5′-flanking region identified two candidate DREs, one at 8138 and one at 3476 nt upstream of the transcription start site. A fragment containing 7073 nt of the SULT1E1 5′-flanking region was ligated into a luciferase reporter plasmid, and this plasmid was used for stable or transient transfection of MCF10A cells. Treatment of stably transfected, preconfluent MCF10A cells with MNF caused a concentration-dependent increase in reporter gene expression (Fig. 7A). In addition, luciferase expression was significantly higher in confluent than in preconfluent stably transfected MCF10A cells (Fig. 7B), indicating that the information necessary for achieving both confluence- and MNF-inducible SULT1E1 transcription is contained within the 7-kb 5′-flanking region. Because this region of the SULT1E1 gene contains only the predicted DRE at −3476, the importance of the −3476 DRE for conferring AhR-mediated regulation to the 7-kb fragment was evaluated. MNF treatment increased luciferase expression in preconfluent MCF10A cells that were transiently transfected with the 7-kb SULT1E1 reporter plasmid, but transfection of a SULT1E1 plasmid containing a site-directed mutation in a core nucleotide of the −3476 DRE did not attenuate MNF-mediated reporter induction (Fig. 7C). In addition, transient transfection of the 7-kb SULT1E1 reporter plasmid into the MCF10A cells that had been engineered for conditional AhR knockdown resulted in doxycycline-inducible reporter expression, and mutation of the DRE did not affect this up-regulation (Fig. 7D). These findings indicate that AhR inhibition/suppression-mediated SULT1E1 up-regulation is not mediated through the DRE at −3476.
Fig. 7.
Effect of MNF treatment, cell confluence, and AhR knockdown on luciferase expression from a reporter plasmid containing 7073 nt of the SULT1E1 5′-flanking sequence. A, MCF10A cells were stably transfected with the SULT1E1-luciferase reporter plasmid, and preconfluent cells were treated with 1 to 10 μM MNF for 24 h, after which they were harvested for measurement of firefly luciferase activity and protein content. Each bar represents the mean relative luciferase activity (normalized to cellular protein content) ± S.D. (three wells per treatment group). Groups that do not share a capital letter are significantly different from each other (p < 0.001). B, preconfluent and confluent MCF10A cells stably transfected with the SULT1E1-luciferase reporter were harvested for measurement of luciferase activity as described in A. ***, p < 0.001 compared with preconfluent cells. C, preconfluent MCF10A cells were transiently transfected with the SULT1E1-luciferase reporter containing either an intact (□) or mutated (■) predicted DRE at −3476, treated for 24 h with 1 to 10 μM MNF, and harvested for measurement of luciferase activities. Each bar represents the mean relative luciferase activity (firefly/Renilla) ± S.D. (three wells per treatment group). Groups that do not share a capital letter are significantly different from each other (p < 0.05). D, preconfluent MCF10A cells engineered for conditional knockdown of AhR were treated with either 0.1% DMSO or 1 μg/ml doxycycline for 96 h and transiently transfected with SULT1E1-luciferase reporter containing intact or mutant DRE. At 24 h after transfection, cells were harvested for measurement of firefly and Renilla luciferase activities. Groups that do not share a capital letter are significantly different from each other (p < 0.001).
Discussion
The impact of manipulations that alter cell-cell or cell-matrix contacts on AhR target gene expression was first investigated by Sadek and Allen-Hoffmann (1994a), who reported that suspension of cultured human keratinocytes caused increased expression of CYP1A1 and other AhR target genes. Further studies using Hepa1c1c7 murine hepatoma cells and variants defective in AhR signaling confirmed that cell suspension caused activation of the AhR (Sadek and Allen-Hoffmann, 1994b). Subsequently, Monk et al. (2001) reported that suspension of cultured rat keratinocytes caused transient AhR activation and CYP1A1 induction and that cotreatment with the AhR antagonist, α-naphthoflavone, inhibited suspension-mediated CYP1A1 induction.
Cho et al. (2004) then demonstrated that either suspension or monolayer culture at low confluence caused AhR activation in C3H10T1/2 fibroblast clonal sublines. An important conclusion from these studies was that disruption of cell-cell contact was responsible for AhR activation. Relative to confluent C3H10T1/2 cells, in ∼70% confluent cells, there was ∼4-fold activation of an AhR-responsive reporter, which is approximately the same magnitude of CYP1A1 mRNA and pGudLuc1.1 up-regulation that we observed in confluent versus preconfluent MCF10A cells (Cho et al., 2004). Treatments with several inhibitors of processes involved in AhR · ARNT complex formation inhibited AhR activation whether it was produced by TCDD treatment or loss of cell-cell contact, indicating that these stimuli induced AhR · ARNT complex formation through the same mechanism. However, some treatments that interfere with the transcriptional activity of AhR · ARNT complexes produced stimulus-dependent effects on AhR function, suggesting that the AhR · ARNT complexes are regulated differently after TCDD treatment and loss of cell-cell contact. A notable difference from the findings of Monk et al. (2001) was that α-naphthoflavone treatment blocked TCDD-mediated AhR activation but not activation by loss of cell-cell contact. Consistent with Monk et al., we found that AhR inhibitor treatment reduced the level of CYP1A1 expression in preconfluent MCF10A cells.
In addition, Ikuta et al. (2004) reported that cell density influenced the subcellular distribution of the AhR. The AhR was predominantly nuclear in the HaCaT human keratinocyte cell line at low confluence, both nuclear and cytoplasmic at preconfluence, and predominantly cytoplasmic at confluence. They also used a cell-scrape model of wound healing to demonstrate that the AhR became activated in the loosely associated cells at the border of the wound margin (Ikuta et al., 2004). These investigators hypothesized that loss of cell-cell contact activates signaling events, possibly mediated through p38 mitogen-activated protein kinase, which increase the phosphorylation of the AhR at its nuclear export signal, causing AhR to accumulate in the nucleus (Ikuta et al., 2004).
AhR function has also been linked to the cell cycle. For example, Santini et al. (2001) used centrifugal elutriation to isolate populations of TCDD-treated human monocytic U937 cells in different phases of the cell cycle and reported that late G1/early S phase cells had CYP1A1 mRNA contents that were ∼1.4- and 3-fold higher than the contents of asynchronous/early G1 and G2/M cultures, respectively. These studies suggest that the transcriptional activation of AhR target genes by TCDD is cell cycle-dependent and suppressed in G2/M cells. However, both Cho et al. (2004) and Ikuta et al. (2004) reported that low cell density-mediated AhR activation was not dependent on cell cycle phase.
One possible mechanism for AhR-mediated transcriptional suppression is through the binding of the activated AhR · ARNT complex to an inhibitory DRE. Safe and Wormke (2003) reported that certain genes (i.e., cathepsin D, c-fos, pS2, and Hsp27) contain pentanucleotide GCGTG sites that correspond to the core DRE motif and function as inhibitory DREs in that the binding of liganded AhR to these sites inhibits estrogen-mediated transcriptional activation by disrupting the binding of ER or other transcription factors to activating sites that are located in proximity to the DREs. Computational analysis of 10 kb of the SULT1E1 5′-flanking region identified two high-scoring AhR · ARNT binding sites: one at 8138 and one at 3476 nt upstream of the transcription start site. Of note, both of these DRE sites were identified as matches to the V$AHRARNT.03 matrix, which was compiled using, among other sequences, the inhibitory DRE sites contained in the cathepsin D and Hsp27 genes. Although a reporter construct containing ∼7 kb of SULT1E1 5′-flanking sequence and therefore the DRE at −3476 showed significant up-regulation in response to MNF treatment, AhR knockdown, or cell confluence, site-directed mutagenesis of the DRE did not affect the up-regulation, suggesting that this DRE does not play a role in the negative regulation of SULT1E1 transcription.
It is therefore probable that the AhR suppresses SULT1E1 transcription by modulating the activity of some other transcription factor. The AhR has been shown to interact physically with a variety of transcription factors or transcription factor modulatory proteins, including nuclear receptors ERα, COUP-TFI, and ERRα1 (Klinge et al., 2000), nuclear factor-κB subunits RelA and RelB (Tian et al., 1999; Vogel et al., 2007), the cell cycle regulatory protein Rb (Ge and Elferink, 1998), and the apoptosis regulatory transcription factor E2F1 (Puga et al., 2009), thereby modulating their activities either positively or negatively. The AhR also engages in cross-talk interactions with mitogen-activated protein kinases (Puga et al., 2009).
Our results add to the growing number of mechanisms whereby the AhR modulates estrogenic activity. Antiestrogenic effects of AhR ligands, in particular, have been extensively studied, and several mechanisms underlying such effects have been reported (for review, see Safe and Wormke, 2003), including the following; 1) AhR-mediated induction of enzymes (e.g., CYP1B1) that metabolize estrogens and thereby reduce tissue estrogen concentrations (Takemoto et al., 2004); 2) AhR-mediated induction of a transcription inhibitory factor (Rogers and Denison, 2002); 3) an inhibitory action mediated by the nonproductive binding of liganded AhR to an ER target gene, which prevents ER from binding (Krishnan et al., 1995); 4) AhR-mediated reduction of cellular ER levels by either suppression of ER transcription (Tian et al., 1998) or acceleration of ER degradation (Wormke et al., 2003); and 5) AhR-mediated transcriptional activation of its target genes, resulting in competition for recruitment of the limited pool of coactivators that are shared by the AhR and ER. In this regard, ARNT is said to function as a coactivator for ER, but with selectivity for ERβ. Thus, ARNT recruitment to AhR target genes can reduce the transcription of ER target genes (Rüegg et al., 2008). Concerning proestrogenic effects, Ohtake et al. (2003) reported that AhR ligand treatment can induce ER-mediated transcription through the formation of an AhR · ARNT · ER complex (Brosens and Parker, 2003). In other studies, AhR ligands have been found to activate ER-mediated transcriptional activity without a requirement for the AhR (Abdelrahim et al., 2006; Shipley and Waxman, 2006). Abdelrahim et al. (2006) reported that AhR ligands 3-methylcholanthrene and 3,3′,4,4′,5-pentachlorobiphenyl were capable of activating ERs in MCF7 breast cancer cells, whereas Shipley and Waxman (2006) found that 3-methylcholanthrene but not 3,3′,4,4′,5-pentachlorobiphenyl or TCDD functioned as an ER agonist in Ishikawa uterine cancer cells. In another study, Boverhof et al. (2006) reported that TCDD treatment of ovariectomized mice altered the expression of numerous uterine genes that were comparably regulated by 17α-ethynylestradiol. The AhR was also shown to be required for aromatase expression in mouse ovary, and treatment with the AhR ligand 9,10-dimethyl-1,2-benzanthracene increased ovarian expression of aromatase (Baba et al., 2005). By demonstrating that AhR activation suppresses expression of SULT1E1, a major estrogen-inactivating enzyme, our study provides another mechanism by which AhR can regulate estrogenicity.
Most available data suggest that SULT1E1 expression is decreased in breast cancer cells relative to normal breast epithelial cells (Falany and Falany, 1996; Fu et al., 2010), and SULT1E1 immunoreactivity has been inversely correlated with breast tumor size in patients (Suzuki et al., 2003). However, we have no evidence that loss of cell-cell contact and AhR activation contribute to the decreased SULT1E1 expression that occurs during breast carcinogenesis. We recently reported that low expression of SULT1E1 in MCF10A-derived cancer cell lines can be increased by treatment with trichostatin A (Fu et al., 2010), suggesting that chromatin modifications probably underlie the suppression of SULT1E1 that occurs in breast cancer cells. We suggest that AhR-mediated regulation of SULT1E1 might play an important role in modulating estrogen mitogenicity in normal breast tissue. When the breast cells are in a nonproliferative state, it is essential that the growth stimulatory effects of estrogens be held to an absolute minimum. Therefore, cell-cell contact triggers molecular events that include inhibition of AhR activity and up-regulation of SULT1E1 activity. When the breast cells switch to a proliferative state, a lessening of cell-cell contact causes activation of AhR activity and suppression of SULT1E1 expression, resulting in increased active estrogen levels in the breast microenvironment. Such an effect might be relevant to the proliferation of normal breast epithelial cells, for example, during puberty, or of preneoplastic breast epithelial cells undergoing hyperplasia, but it will be necessary to evaluate this possibility in an in vivo system.
Acknowledgments
We thank Dr. Michael Denison for providing the pGudLuc1.1 reporter plasmid and Dr. John Reiners for assistance with the AhR and ARNT Western blot analysis.
This work was supported by the National Institutes of Health National Institute of Environmental Health Sciences [Grant ES05823]; and Uniting Against Lung Cancer.
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.111.185173.
- SULT
- cytosolic sulfotransferase
- SULT1E1
- estrogen sulfotransferase
- ER
- estrogen receptor
- AhR
- aryl hydrocarbon receptor
- TCDD
- 2,3,7,8-tetrachlorodibenzo-p-dioxin
- MNF
- 3′-methoxy-4′-nitroflavone
- BrU
- 5-bromouridine
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- DMSO
- dimethyl sulfoxide
- ARNT
- aryl hydrocarbon receptor nuclear translocator
- PCR
- polymerase chain reaction
- hnRNA
- heterogeneous nuclear RNA
- RT
- reverse transcriptase
- nt
- nucleotide
- PBS
- phosphate-buffered saline
- DRE
- dioxin response element
- kb
- kilobase
- Ct
- cycle threshold.
Authorship Contributions
Participated in research design: Fu, Ljungman, Kocarek, and Runge-Morris.
Conducted experiments: Fu, Fang, and Paulsen.
Contributed new reagents or analytic tools: Ljungman.
Performed data analysis: Fu, Paulsen, Ljungman, and Kocarek.
Wrote or contributed to the writing of the manuscript: Fu, Ljungman, Kocarek, and Runge-Morris.
References
- Abdelrahim M, Ariazi E, Kim K, Khan S, Barhoumi R, Burghardt R, Liu S, Hill D, Finnell R, Wlodarczyk B, et al. (2006) 3-Methylcholanthrene and other aryl hydrocarbon receptor agonists directly activate estrogen receptor α. Cancer Res 66:2459–2467 [DOI] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. (2008) Regulation of sulfotransferase enzymes by prototypical microsomal enzyme inducers in mice. J Pharmacol Exp Ther 324:612–621 [DOI] [PubMed] [Google Scholar]
- Baba T, Mimura J, Nakamura N, Harada N, Yamamoto M, Morohashi K, Fujii-Kuriyama Y. (2005) Intrinsic function of the aryl hydrocarbon (dioxin) receptor as a key factor in female reproduction. Mol Cell Biol 25:10040–10051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boverhof DR, Kwekel JC, Humes DG, Burgoon LD, Zacharewski TR. (2006) Dioxin induces an estrogen-like, estrogen receptor-dependent gene expression response in the murine uterus. Mol Pharmacol 69:1599–1606 [DOI] [PubMed] [Google Scholar]
- Brosens JJ, Parker MG. (2003) Gene expression: oestrogen receptor hijacked. Nature 423:487–488 [DOI] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. (2005) MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21:2933–2942 [DOI] [PubMed] [Google Scholar]
- Cho YC, Zheng W, Jefcoate CR. (2004) Disruption of cell-cell contact maximally but transiently activates AhR-mediated transcription in 10T1/2 fibroblasts. Toxicol Appl Pharmacol 199:220–238 [DOI] [PubMed] [Google Scholar]
- Cuthill S, Wilhelmsson A, Poellinger L. (1991) Role of the ligand in intracellular receptor function: receptor affinity determines activation in vitro of the latent dioxin receptor to a DNA-binding form. Mol Cell Biol 11:401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falany CN. (1997) Enzymology of human cytosolic sulfotransferases. FASEB J 11:206–216 [DOI] [PubMed] [Google Scholar]
- Falany CN, Krasnykh V, Falany JL. (1995) Bacterial expression and characterization of a cDNA for human liver estrogen sulfotransferase. J Steroid Biochem Mol Biol 52:529–539 [DOI] [PubMed] [Google Scholar]
- Falany JL, Falany CN. (1996) Expression of cytosolic sulfotransferases in normal mammary epithelial cells and breast cancer cell lines. Cancer Res 56:1551–1555 [PubMed] [Google Scholar]
- Fu J, Weise AM, Falany JL, Falany CN, Thibodeau BJ, Miller FR, Kocarek TA, Runge-Morris M. (2010) Expression of estrogenicity genes in a lineage cell culture model of human breast cancer progression. Breast Cancer Res Treat 120:35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison PM, Tullis K, Aarts JM, Brouwer A, Giesy JP, Denison MS. (1996) Species-specific recombinant cell lines as bioassay systems for the detection of 2,3,7,8-tetrachlorodibenzo-p-dioxin-like chemicals. Fundam Appl Toxicol 30:194–203 [DOI] [PubMed] [Google Scholar]
- Ge NL, Elferink CJ. (1998) A direct interaction between the aryl hydrocarbon receptor and retinoblastoma protein. Linking dioxin signaling to the cell cycle. J Biol Chem 273:22708–22713 [DOI] [PubMed] [Google Scholar]
- Glatt H. (2000) Sulfotransferases in the bioactivation of xenobiotics. Chem Biol Interact 129:141–170 [DOI] [PubMed] [Google Scholar]
- Ikuta T, Kobayashi Y, Kawajiri K. (2004) Cell density regulates intracellular localization of aryl hydrocarbon receptor. J Biol Chem 279:19209–19216 [DOI] [PubMed] [Google Scholar]
- Klinge CM, Kaur K, Swanson HI. (2000) The aryl hydrocarbon receptor interacts with estrogen receptor α and orphan receptors COUP-TFI and ERRα1. Arch Biochem Biophys 373:163–174 [DOI] [PubMed] [Google Scholar]
- Krishnan V, Porter W, Santostefano M, Wang X, Safe S. (1995) Molecular mechanism of inhibition of estrogen-induced cathepsin D gene expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in MCF-7 cells. Mol Cell Biol 15:6710–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk SA, Denison MS, Rice RH. (2001) Transient expression of CYP1A1 in rat epithelial cells cultured in suspension. Arch Biochem Biophys 393:154–162 [DOI] [PubMed] [Google Scholar]
- Ohtake F, Takeyama K, Matsumoto T, Kitagawa H, Yamamoto Y, Nohara K, Tohyama C, Krust A, Mimura J, Chambon P, et al. (2003) Modulation of oestrogen receptor signalling by association with the activated dioxin receptor. Nature 423:545–550 [DOI] [PubMed] [Google Scholar]
- Puga A, Ma C, Marlowe JL. (2009) The aryl hydrocarbon receptor cross-talks with multiple signal transduction pathways. Biochem Pharmacol 77:713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga A, Maier A, Medvedovic M. (2000) The transcriptional signature of dioxin in human hepatoma HepG2 cells. Biochem Pharmacol 60:1129–1142 [DOI] [PubMed] [Google Scholar]
- Qian YM, Sun XJ, Tong MH, Li XP, Richa J, Song WC. (2001) Targeted disruption of the mouse estrogen sulfotransferase gene reveals a role of estrogen metabolism in intracrine and paracrine estrogen regulation. Endocrinology 142:5342–5350 [DOI] [PubMed] [Google Scholar]
- Rivera SP, Saarikoski ST, Hankinson O. (2002) Identification of a novel dioxin-inducible cytochrome P450. Mol Pharmacol 61:255–259 [DOI] [PubMed] [Google Scholar]
- Rogers JM, Denison MS. (2002) Analysis of the antiestrogenic activity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in human ovarian carcinoma BG-1 cells. Mol Pharmacol 61:1393–1403 [DOI] [PubMed] [Google Scholar]
- Rüegg J, Swedenborg E, Wahlström D, Escande A, Balaguer P, Pettersson K, Pongratz I. (2008) The transcription factor aryl hydrocarbon receptor nuclear translocator functions as an estrogen receptor β-selective coactivator, and its recruitment to alternative pathways mediates antiestrogenic effects of dioxin. Mol Endocrinol 22:304–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge-Morris M. (1998) Regulation of sulfotransferase gene expression by glucocorticoid hormones and xenobiotics in primary rat hepatocyte culture. Chem Biol Interact 109:315–327 [DOI] [PubMed] [Google Scholar]
- Runge-Morris M, Wilusz J. (1994) Suppression of hydroxysteroid sulfotransferase-a gene expression by 3-methylcholanthrene. Toxicol Appl Pharmacol 125:133–141 [DOI] [PubMed] [Google Scholar]
- Sadek CM, Allen-Hoffmann BL. (1994a) Cytochrome P450IA1 is rapidly induced in normal human keratinocytes in the absence of xenobiotics. J Biol Chem 269:16067–16074 [PubMed] [Google Scholar]
- Sadek CM, Allen-Hoffmann BL. (1994b) Suspension-mediated induction of Hepa 1c1c7 Cyp1a-1 expression is dependent on the Ah receptor signal transduction pathway. J Biol Chem 269:31505–31509 [PubMed] [Google Scholar]
- Safe S, Wormke M. (2003) Inhibitory aryl hydrocarbon receptor-estrogen receptor α cross-talk and mechanisms of action. Chem Res Toxicol 16:807–816 [DOI] [PubMed] [Google Scholar]
- Santini RP, Myrand S, Elferink C, Reiners JJ., Jr (2001) Regulation of Cyp1a1 induction by dioxin as a function of cell cycle phase. J Pharmacol Exp Ther 299:718–728 [PubMed] [Google Scholar]
- Shipley JM, Waxman DJ. (2006) Aryl hydrocarbon receptor-independent activation of estrogen receptor-dependent transcription by 3-methylcholanthrene. Toxicol Appl Pharmacol 213:87–97 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Nakata T, Miki Y, Kaneko C, Moriya T, Ishida T, Akinaga S, Hirakawa H, Kimura M, Sasano H. (2003) Estrogen sulfotransferase and steroid sulfatase in human breast carcinoma. Cancer Res 63:2762–2770 [PubMed] [Google Scholar]
- Takemoto K, Nakajima M, Fujiki Y, Katoh M, Gonzalez FJ, Yokoi T. (2004) Role of the aryl hydrocarbon receptor and Cyp1b1 in the antiestrogenic activity of 2,3,7,8-tetrachlorodibenzo-p-dioxin. Arch Toxicol 78:309–315 [DOI] [PubMed] [Google Scholar]
- Thomas RD, Green MR, Wilson C, Weckle AL, Duanmu Z, Kocarek TA, Runge-Morris M. (2006) Cytochrome P450 expression and metabolic activation of cooked food mutagen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in MCF10A breast epithelial cells. Chem Biol Interact 160:204–216 [DOI] [PubMed] [Google Scholar]
- Tian Y, Ke S, Denison MS, Rabson AB, Gallo MA. (1999) Ah receptor and NF-κB interactions, a potential mechanism for dioxin toxicity. J Biol Chem 274:510–515 [DOI] [PubMed] [Google Scholar]
- Tian Y, Ke S, Thomas T, Meeker RJ, Gallo MA. (1998) Transcriptional suppression of estrogen receptor gene expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). J Steroid Biochem Mol Biol 67:17–24 [DOI] [PubMed] [Google Scholar]
- Vogel CF, Sciullo E, Matsumura F. (2007) Involvement of RelB in aryl hydrocarbon receptor-mediated induction of chemokines. Biochem Biophys Res Commun 363:722–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormke M, Stoner M, Saville B, Walker K, Abdelrahim M, Burghardt R, Safe S. (2003) The aryl hydrocarbon receptor mediates degradation of estrogen receptor α through activation of proteasomes. Mol Cell Biol 23:1843–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]