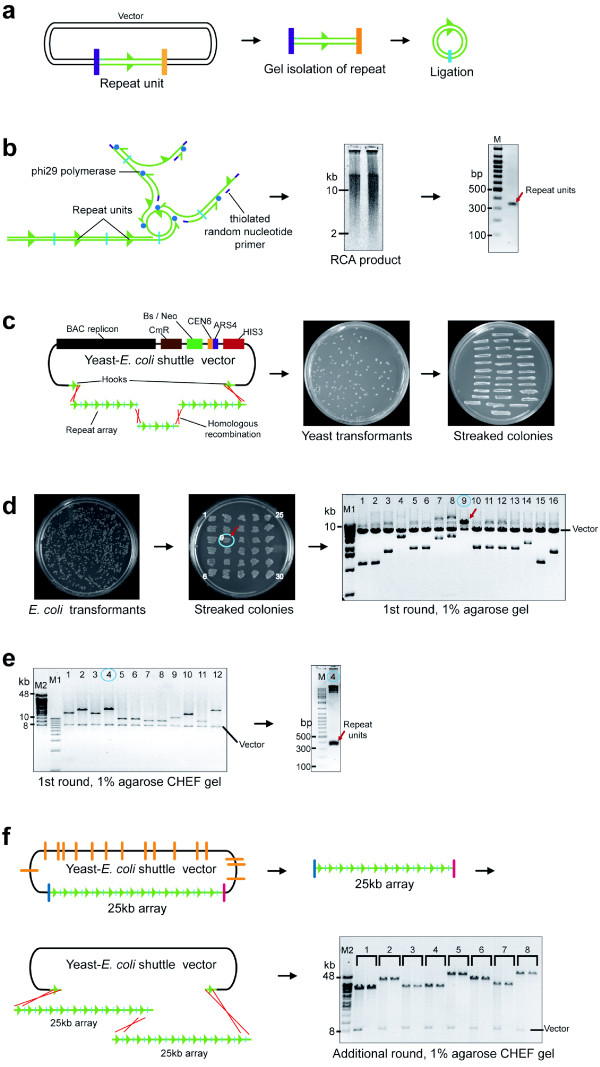
Figure 1.
Scheme of the repeats assembly into synthetic DNA arrays. (a) The DNA repeat isolated from the vector (in purple and orange) is ligated to make a circular molecule. (b) The RCA products were generated from a 340 bp alphoid dimer. Cleavage of RCA products with an enzyme results in restoration of the input repeat. (c) Recombinational assembly includes co-transformation of RCA products into yeast along with a TAR vector (YAC/BAC) containing repeat-specific targeting hooks. End-to-end recombination of DNA fragments, followed by interaction of the recombined fragments with the vector hooks, results in the rescue of arrays as circular YACs. His+ transformants and pooled colonies are shown. (d) Transferring of YACs into bacterial cells. E. coli transformants and streaked colonies are shown. BAC DNAs from randomly picked up colonies were restricted by an endonuclease that releases the vector part (7 kb) and arrays. The size of arrays varies from 2 to 12 kb. BAC DNA from colony #9 with the largest array is marked by the red arrow. (e) CHEF analysis of BACs with the largest arrays chosen after screening 60 E. coli transformants. The size of the inserts varies from 9 to 25 kb. The tandem repeat structure of one array (clone #4 with the size ~25 kb) is confirmed by EcoRI digestion (f) An additional round of recombinational assembly to further increase the size of the array. Representative CHEF analysis of 8 BACs is shown. Restriction of BAC DNAs was done by an endonuclease that cleaves the molecule at insert/vector junctions (arrays are between 40 and 60 kb) and by double digestion with an additional endonuclease that cuts the vector part completely.