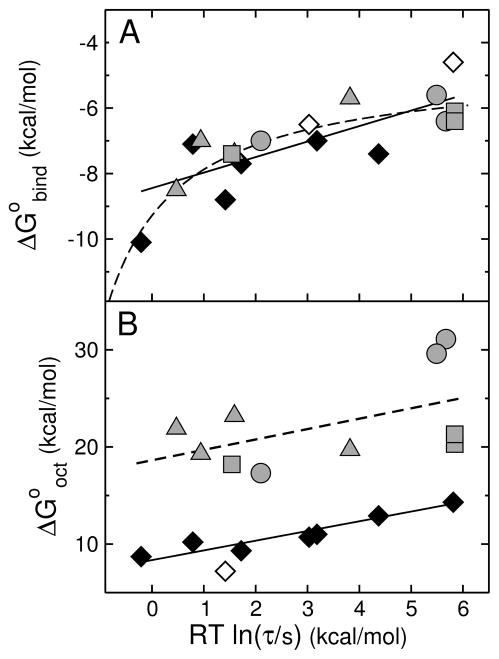
FIGURE 9.
(A) Gibbs energy of binding to the membrane interface determined experimentally ( ) as a function of the mean characteristic time τ for CF release from POPC LUVs. Each gray symbol corresponds to a peptide examined here: δ-lysin, DL-1, DL-2a, and DL-2b are shown by gray triangles; cecropin A, CE-1, and CE-2, by gray circles; and magainin 2, MG-1, and MG-2, by gray squares. The data for TP10 variants previously published (16) are shown here for comparison (diamonds): TP10W, TPW-1, TPW-2, and TPW-3; TP10, TP10-COO−, TP10W-COO−, and TP10-7MC (38, 58). The black symbols correspond to experimental data; the open symbols correspond to TP10 and TP10-COO−, for which τ is experimental but the binding affinity is calculated with the Wimley-White scale (neither contains Trp). The straight line is a fit, with a slope of 0.5. The dashed line represents qualitatively the expected behavior limited by diffusion or bilayer response. (B) Gibbs energy of transfer to octanol calculated with the Wimley-White scale ( ) as a function of the mean characteristic time τ for CF release from POPC LUV. The points correspond to the same peptides as in (A) and the same symbols were used. Again the TP10 variant data are from our previous paper (16). The open circle corresponds to TP10-7MC for which was estimated assuming Tyr for the Lys-MC residue (16). The lines are fits, which have slopes of 1.