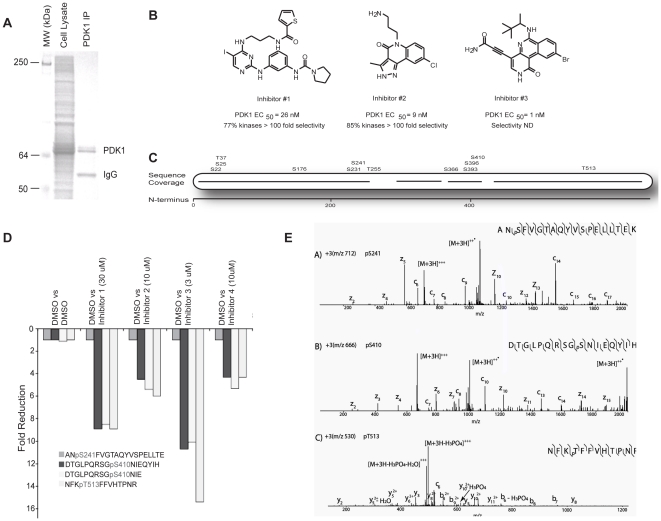
Figure 4. Relative quantification of hPDK1 phosphorylation from 293T cells treated with ATP competitive PDK1 inhibitors using SILAC-IAP-MS.
A) SDS-PAGE and isolation of the PDK1 protein band for LC-MS/MS analysis. B) PDK1 tool compounds representing three structurally distinct chemical classes. PDK1 enzymatic activity is shown (EC50). C) Mapping of phosphorylation sites of immunoprecipitated PDK1 from 293T cells identifies 12 Ser/Thr phosphorylation sites with ∼95% sequence coverage. D) Relative quantification by SILAC-IAP-MS showed a greater than 4 fold reduction of two phosphorylation sites (pS410, and pT513) in samples treated with PDK1 inhibitors compared to samples treated with DMSO. In contrast, no change in phosphorylation was detected in the self-to-self control sample where 293T cells grown in heavy and light media were both treated with DMSO prior to mixing. E) MS/MS spectra for pS241 (ANpSFVGTAQYVSPELLTEK), pS410 (DTGLPQRSGpSNIEQYIH), and pT513 (NFKpTFFVHTPNR) containing peptides.