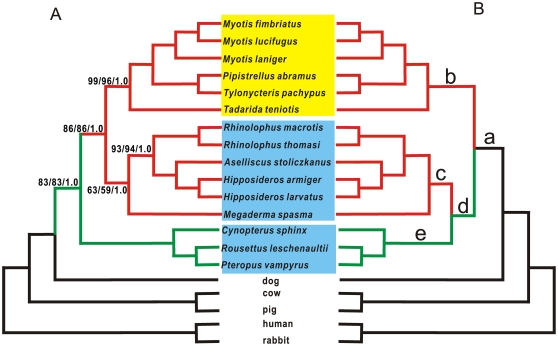
Figure 1.
(A) Putative phylogeny based on amino acid sequences of KCNQ4. Values on the branch indicate support from maximum parsimony (MP), maximum likelihood (ML), and Bayesian inference (BI), respectively. The green and red branches indicate echolocating and non-echolocating bats, respectively. The yellow and blue boxes show bats in the suborders of Yangochiroptera and Yinpterochiroptera, respectively. (B) Putative gene tree for KCNQ4 nucleotide sequences using MP and ML. The topology is consistent with that of species tree and bootstrap values are not shown. The letters on the different branches indicate the targets in our selection tests.