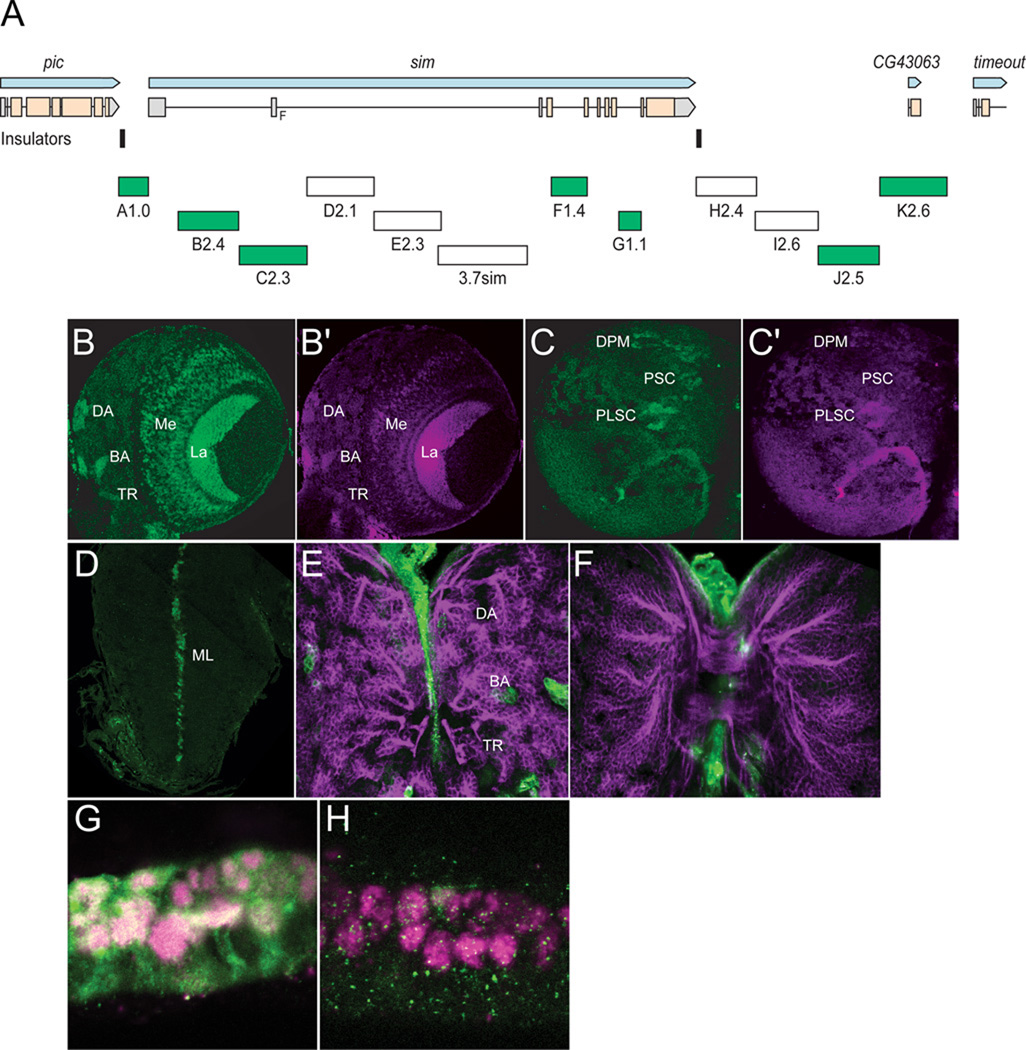
Fig. 3. Transgenic analysis of the sim regulatory region.
(A) Schematic of a 37.8 kb genomic region that includes the sim gene and neighboring pic CG43063, and timeout genes. Two insulator protein binding sites surrounding sim are indicated by vertical lines. Fragments that were analyzed by Gal4 transgenesis are labeled A–K and include the fragment size in kb. The 3.7sim fragment has been previously described (Nambu et al., 1991). Green boxes indicate fragments that drive postembryonic expression, while fragments with no postembryonic expression are unfilled. (B,B’) A1.0-GFP.nls 3rd instar larval brains were stained with anti-GFP (green) and anti-Sim (magenta) to assess whether GFP co-localizes with Sim+ brain cells. On the anterior side of the brain, A1.0-GFP.nls drives expression in DAMv1/2 (DA), BAmas1/2 (BA), and TRdm (TR). In addition, GFP expression is detected in two optic lobe ganglia, the lamina (La) and medulla (Me). (C,C’) On the posterior side of the brain, A1.0-GFP.nls drives GFP expression in DPMm1–3 neurons and DPMpm2 neurons (DPM), PLSC neurons, and PSC neurons. (D) A1.0-GFP.nls drives GFP expression in the midline cells (ML) of the larval ventral nerve cord cells. (E,F) Mutant version of A1.0-GFP.nls, in which both Sim:Tgo binding sites were mutated, was analyzed. The larval brain was stained for GFP (green) and MAb BP106 (magenta). (E) On the anterior side of the brain, the BP106 staining indicates the location of the DAMv1/2 (DA), BAmas1/2 (BA), and TRdm (TR) neurons, and these cells were GFP−. (F) On the posterior side of the same brain, there was an absence of GFP in brain neurons. (G,H) Sagittal views of stage 11 A1.0-GFP.nls embryos with (G) unmutated A1.0-GFP.nls and (H) a version of A1.0-GFP.nls with the Sim:Tgo sites mutated. Single segment is shown with internal up and anterior to the left. Embryos are stained for GFP (green) and anti-Sim (magenta), which stains all midline cells. The unmutated A1.0-GFP.nls drives robust expression in all midline cells, whereas expression is absent in the mutated version.