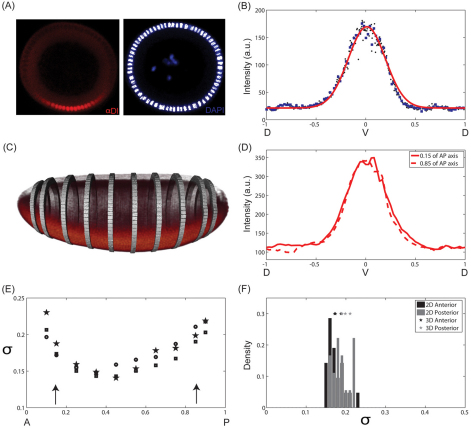
Fig. 1.
Data collection. (A) End-on imaging: DV cross-section of a Drosophila embryo stained using antibody against Dl (left) and DAPI (right). Ventral side is at the bottom and dorsal side is at the top. The nuclear mask (white) was established based on the DAPI signal. (B) Representative spatial pattern of nuclear Dl (black dots) as a function of DV position. The gradient is interpolated onto a uniform grid (blue squares). As the nuclei are spaced very tightly, linear interpolation does not introduce any artifact to the gradient. Solid red line represents the Gaussian fit that is used to find the ventralmost position along the DV axis. (C) 3D imaging of a Drosophila embryo stained with α-Dl antibody using scanned light sheet-based microscopy. The bands correspond to positions along the AP axis of the embryo where the DV pattern of nuclear Dl was quantified. (D) DV profile of nuclear Dl at 15% (solid) and 85% (dashed) of the AP axis from the anterior pole in one embryo. (E) Gaussian width (σ) of nuclear Dl profile as a function of position along the AP axis for three independent embryos. Arrows indicate the depth at which cross-sectional views are imaged using the microfluidic device. (F) Comparison of the estimated Gaussian widths of nuclear Dl profiles, measured 15% from anterior and posterior poles, obtained by scanned light sheet-based microscopy (3D) with those obtained by end-on imaging in a microfluidic device (2D).