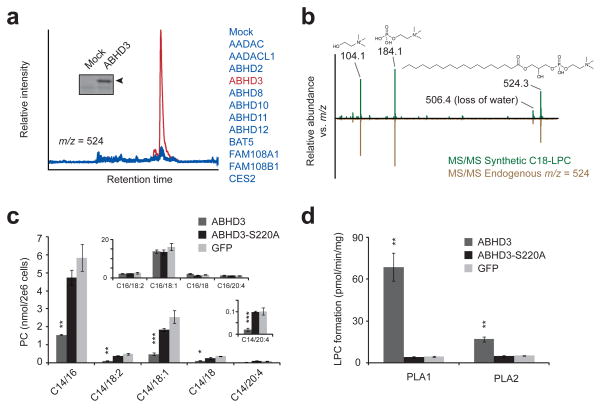
Fig. 1.
Metabolomic profiling of an enzyme library identifies metabolites altered by ABHD3 overexpression. (a) Representative overlaid extracted ion chromatograms at m/z = 523.5–524.5 from HEK293T cells transfected with ABHD3 (red trace) versus other enzymes (blue traces). Insert: Activity-based labeling using the serine hydrolase-directed probe fluorophosphonate rhodamine (FP-Rh) in mock versus ABHD3-transfected cells. Arrowhead designates FP-Rh-labeled ABHD3 protein. (b) Representative MS/MS fragmentation of synthetic C18-LPC (top, in green) and endogenous m/z = 524 (bottom, in brown) gave identical daughter ions of 104.1 (choline), 184.1 (phosphocholine), and 506.1 (dehydro-C18-LPC). (c) Targeted MRM measurements of phosphocholines (PCs) from C8161 cells stably overexpressing epitope-tagged ABHD3 (dark grey), the catalytically dead ABHD3-S220A mutant (black), or GFP (light grey). PC species are indicated by C#/##, where the # indicates the sn-1 acyl chain and ## indicates the sn-2 acyl chain. Top insert: C16-containing PC species; bottom insert: an enlarged graph showing C14/20:4-PC. (d) PLA1 and PLA2 hydrolysis activities for ABHD3 using C14/18:2-PC as a substrate and lysates from stably transfected C8161 cells as the protein source. Data are presented as mean ± standard error; n = 4/group; * P < 0.05, ** P < 0.01, *** P < 0.001 for ABHD3 versus ABHD3-S220A groups.