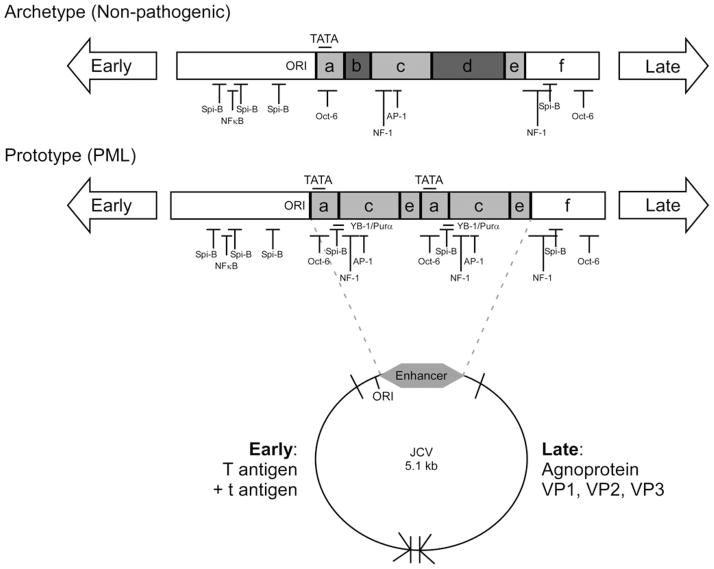
Fig. 2.
Host transcription factor binding sites present in JC virus archetype and prototype regulatory region sequences. The circular map represents the 5.1-kb circular double-stranded DNA genome of JC virus. Early (large T antigen and small t antigen) and late genes (agnoprotein, VP1, VP2, and VP3) are separated by the noncoding viral regulatory region that contains the origin of DNA replication (ORI) and enhancer sequences (gray). The noncoding regulatory regions from the nonpathogenic archetype and PML-associated prototype variants of JC virus are represented in the late orientation for transcription. The origin of DNA replication (ORI), enhancer regions (gray), TATA boxes, and transcription factor binding sites are labeled on each regulatory region diagram. The following cellular transcription factor binding sites are represented: AP-1, NF-1, NFκB, Oct-6, Spi-B, YB-1/Purα