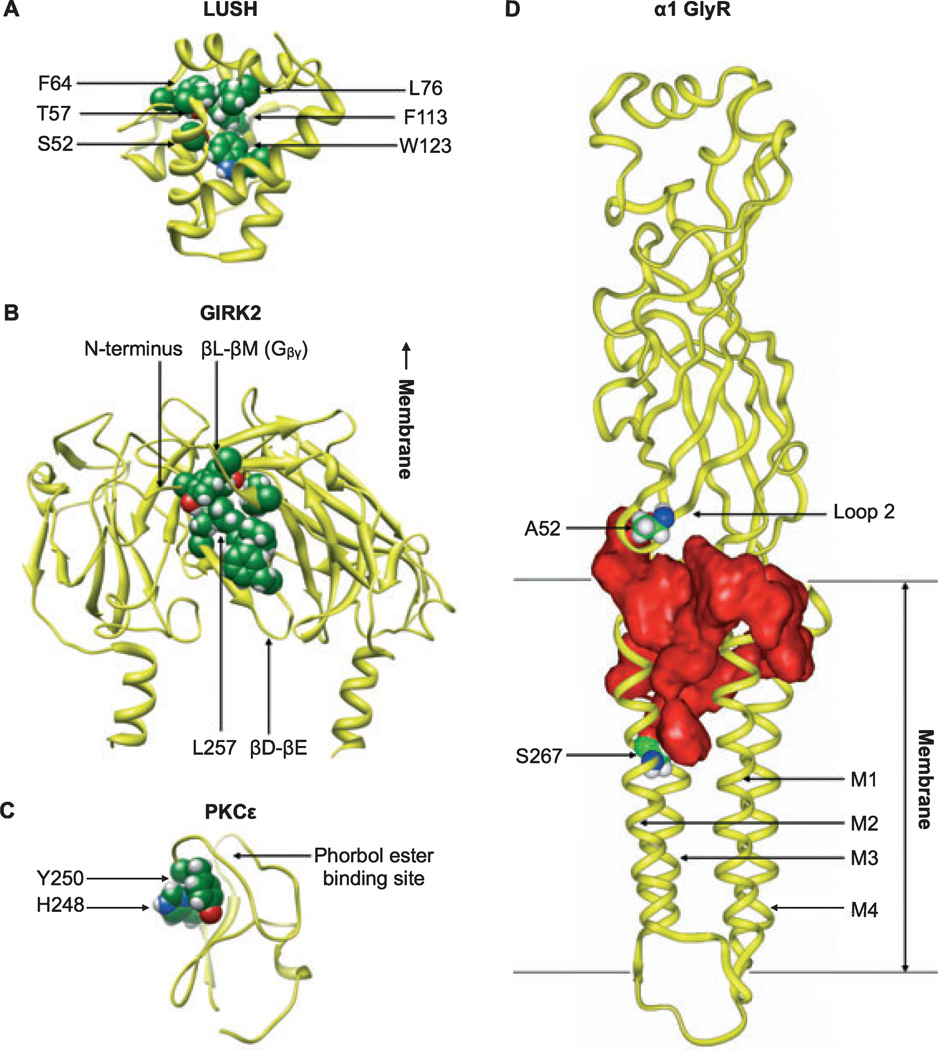
Fig. 1.
Sites of alcohol binding to various protein targets. (A) Coordination of n-alcohols (ethanol, n-propanol, or n-butanol) in the odorant-binding protein LUSH (PDB ID 1OOH) includes hydrogen bonding of S52 and T57 with the alcohol hydroxyl group and hydrophobic interactions of F64, L76, F113, and W123 with the alkyl chain. Residues interacting with n-butanol in the crystal structure are shown as spheres. (B) Binding of alcohol to the GIRK2 intracellular domain (PDB ID 2E4F) may involve rearrangement of residues in the N-terminal domain, the βD-βE loop (including the critical alcohol-binding residue L257), and the βL- βM loop (including sites involved in Gβγ binding). Residues interacting with MPD in the IRK1 crystal structure are mapped onto GIRK2 and shown as spheres. For clarity, only the 2 proximal subunits of the tetramer are shown. (C) Amino acid residues in the C1B domain of PKCε (molecular model based on PKCδ, PDB ID 1PTQ) shown by photolabeling and mass spectrometry to interact with alcohol include H248 and Y250, shown as spheres. Residues in the same groove interact with cyclopropanemethanol in a PKCδ co-crystal structure. (D) A large water-filled cavity (red) contained within a single subunit of the homopentameric α1 GlyR (modeled after the nicotinic acetylcholine ˚ receptor, PDB ID 2BG9) is implicated in ethanol binding. Alcohol-binding residues A52 (Loop 2) and S267 (M2), shown as spheres, are separated by ~28 Å but border a contiguous pocket.