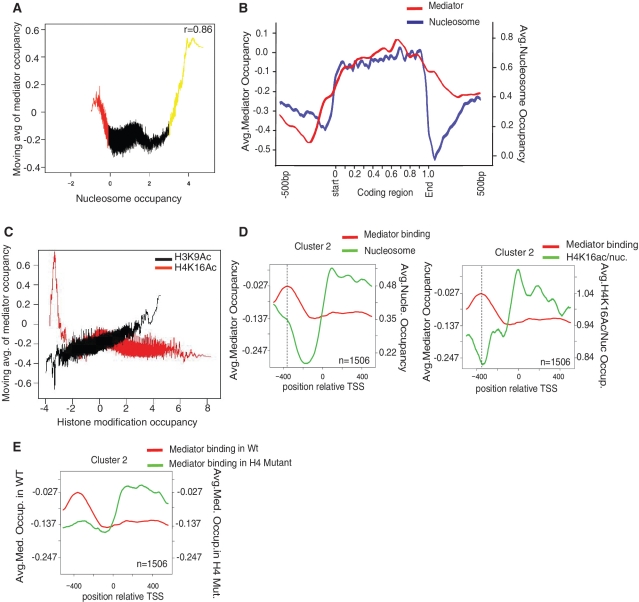
Figure 6.
H4K16 acetylation influences Mediator occupancy in vivo. (A) Moving average analysis of Mediator versus nucleosome occupancy in vivo. Values on the X- and Y-axes are log2 scale. Window size is 150. A correlation coefficient (r = 0.86) is given for the yellow part of the graph (nucleosome occupancy >3). (B) The average Mediator and nucleosome occupancy display similar patterns on highly transcribed genes. The positions relative to the coding regions are shown on the X-axis (with 500 bp of intergenic region on each side being included). Since the genes have different lengths, relative positions are used inside the coding regions. Values on the Y-axis are log2 scale. (C) Moving average analysis of Mediator occupancy versus H4K16ac or H4K9ac occupancy in vivo. The X-axis (log2) display H4K16ac or H3K9ac density, and the Y-axis (log2) correspond to the moving average of Mediator occupancy with a window size of 150. (D) Mediator binding to S. cerevisiae genes were analyzed by k-means clustering (k = 5), and a cluster with high levels of Mediator occupancy at the promoter (1506 genes) was chosen for further analysis. Average levels of Mediator, nucleosome, and H4K16 acetylation (normalized to nucleosome levels) are indicated on the Y-axis (log2). Positions near the TSS (−500 to +500) are indicated on the X-axis. (E) The same analysis as in panel (D), but Mediator occupancy was analyzed both in a wild-type strain and in a histone H4 K8Q, K16Q mutant strain. In these experiments the wild-type and histone mutant Mediator ChIP were performed using a Med18(Srb5)-Flag tagged strain and the anti-Flag antibody (F1804, Sigma).