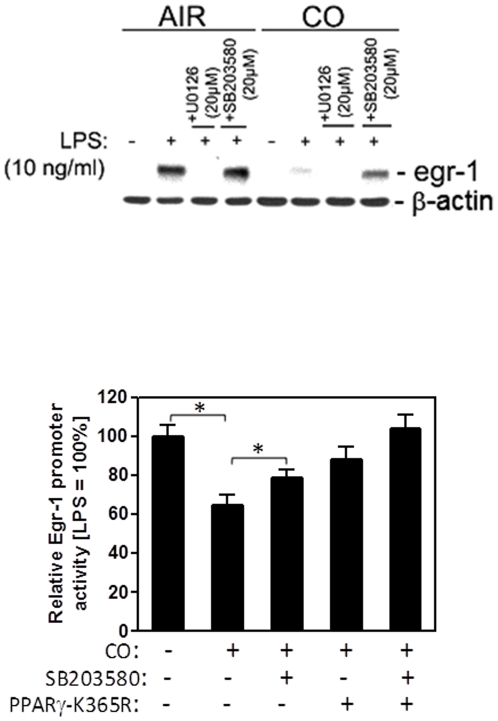
Figure 3. CO-mediated PPARγ-SUMOylation and p38MAPK activity cooperate for full suppression of LPS-induced Egr-1 promoter activity.
(A) RAW 264.7 cells were pretreated with 250 ppm CO or Air for 3 hours, followed by a further 30 min incubation with MAPK inhibitors for p38 (SB203580) or ERK1/2 (U0126) prior to stimulation with LPS (10 ng/ml) for 1 hour. Egr-1 protein expression was analyzed by western blotting and β-actin was used as a loading control. The western blot is representative of three independent experiments. (B) RAW 264.7 cells were transiently transfected with PPARγ-K365R or pcDNA3.1 control vector along with pGL3-Egr1 and phRL-Tk. Thereafter, RAW 264.7 cells were pretreated with 250 ppm CO or Air for 3 hours, followed by a further 30 min incubation with SB203580 or vehicle (DMSO) prior to stimulation with LPS (10 ng/ml). Total proteins were harvested at 6 hr postinduction and dual luciferase activities determined. The value of firefly luciferase was normalized by the rennila luciferase to generate the relative luciferase activity. Bars represent mean values ± SEM of three independent experiments (*P<0.05).