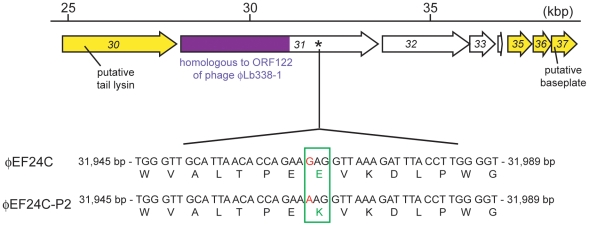
Figure 3. Mutation site in orf31 of phage ΦEF24C-P2 and the features of ORF31 and neighboring ORFs.
The mutation site is shown in the DNA alignments between phage ΦEF24C and phage ΦEF24C-P2, in the bottom row. A point mutation (i.e., substitution) was found on the putative gene orf31, where the nucleotides are shown in red. The point mutation changed an amino acid from glutamic acid (negatively charged amino acid; wild-type phage ΦEF24C) to lysine (positively charged amino acid; mutant phage ΦEF24C-P2), which is colored and boxed in green. The partial genomic map of phage ΦEF24C is shown in the top row. The arrows indicate each ORF. The arrows in yellow indicate ORFs with similarity to other phage proteins. The region colored in purple, the N-terminal region of the ORF31, is similar to the N-terminal region of ORF122 of Lactobacillus paracasei phage ΦLb338-1, with an E-value of 1.0×10−114 in the BLASTP analysis. It occupied 53% of the ORF31 protein sequence. A mutation site is denoted by an asterisk around the C-terminal region of the ORF31, but no protein signature was found. ORF32–34 did not show similarity to the other proteins.