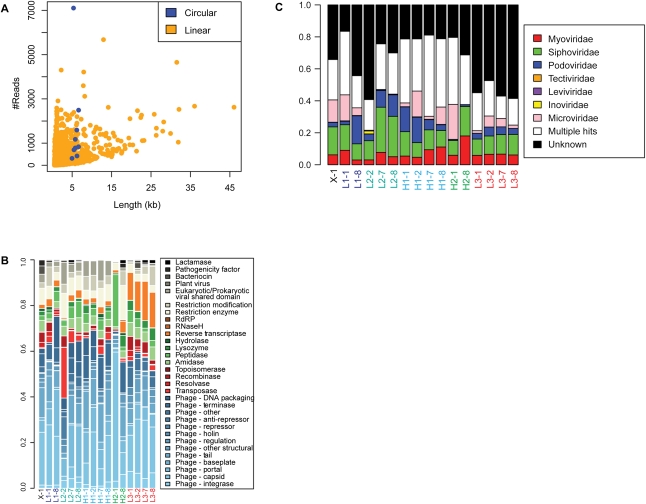
Figure 2.
Assembly and functional annotation of shotgun metagenomic sequences from the human gut virome. (A) Analysis of recruitment of VLP sequence reads into contigs. The y-axis shows the number of sequence reads, the x-axis shows contig length. Pyrosequencing data from the human virome were assembled into 7147 contigs up to 47.8 kb in length. Linear contigs are shown in yellow, circular contigs are shown in blue. (B) Analysis of protein functions in VLP contigs. The functions encoded in VLP contigs were predicted using the Pfam database, then grouped using custom database-relating Pfam domain identifiers to phage functions (Supplemental Table S4). The relative proportions of pyrosequencing reads falling within ORFs of different annotations were plotted according to their sample of origin (y-axis). Each bar is indicated by the sample code, where L or H indicates low- or high-fat diet, the adjacent number indicates the subject number, and the number after the hyphen indicates the day of the study. “X-1” indicates ad-lib diet. (C) Taxonomic classification of VLP communities is consistent across samples. Samples (in columns, labeled as in Figs. 1, 5) are characterized according to the number of sequences from each sample that are assembled into a contig that is classified by taxonomic family. Phage families are indicated by the color code to the right. “Unknown” (black) indicates contigs that cannot be classified in any way. “Multiple hits” (white) indicate contigs that have proteins that are similar to multiple families.