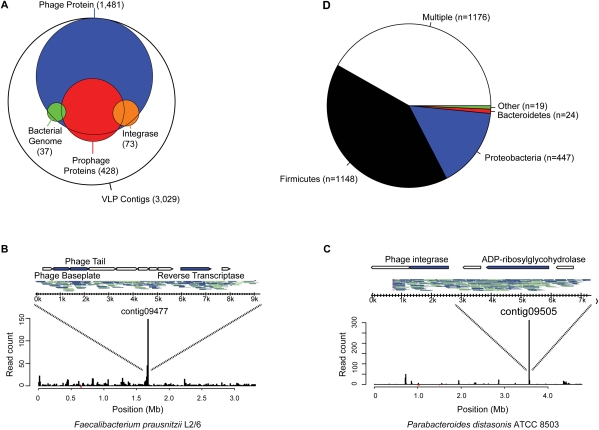
Figure 4.
Analysis of temperate bacteriophages in the human gut virome. (A) Venn diagram indicating frequency of functions associated with temperate phages. VLP contigs were annotated according to the presence of integrase-like sequences (orange), BLAST alignment to sequenced bacterial genomes, suggestive of prophage formation (90% length at 90% identity; green), and presence of multiple genes with significant similarity to a prophage element within the ACLAME database (red). Map of VLP pyrosequencing reads aligning to the genomes of (B) Faecalibacterium prausnitzii L2/6 and (C) Parabacteroides distasonis ATCC 8503 (90% length at 90% identity). (Inset) Contigs that correspond to each peak, the reads that make up each contig, and their annotated genes. Top strand reads are indicated by blue, bottom strand reads by gray. (D) Prevalence of prophage sequences according to bacterial phylum of origin. Contigs with amino acid similarity (E-value < 10−5) to prophage sequences within the ACLAME databases are shown according to which bacterial phylum they match. Any contig with similarity to more than one phylum is classified as “Multiple.” Roughly 39% of VLP contigs (n = 2814) did not have significant similarity to any prophage sequence in this database.