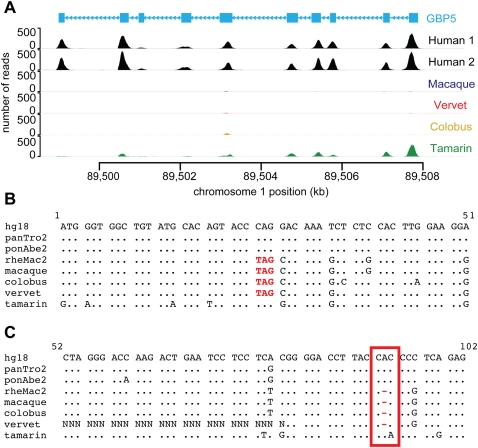
Figure 4.
Examples of gene loss in Old World monkeys. (A) An example of a gene deletion detected by read depth differences between species. The read depth for GBP5 (CCDS722.1) is high in human (Human 1: NA12878 and Human 2: NA18967) and tamarin, but absent in macaque, vervet, and colobus. The absence of GBP5 in the macaque exome sequences is supported by the macaque reference genome. (B) The beginning of the multiple sequence alignment for the gene SNTN (CCDS33779.1) containing a premature stop codon in Old World monkeys. The substitution causing this premature stop codon is high quality (≥Q40) in macaque, vervet, and colobus and confirmed by the macaque reference genome. (C) A portion of the multiple sequence alignment for the gene CCL14 (CCDS32624.1) containing a frameshift in Old World monkeys. (Red box) A one-base gap in macaque, vervet and colobus, which disrupts the reading frame of the latter half of the gene. This gap is surrounded by high-quality (≥Q40) bases and also supported by the macaque reference genome.