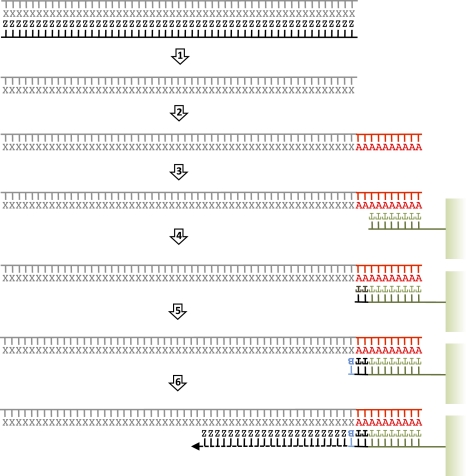
Figure 1.
Helicos tSMS: an overview (adapted from Hart et al. 2010 and reprinted with permission from Elsevier Ltd. © 2010). Ancient DNA molecules are denatured into single strands (step 1), tailed with poly(A) (step 2), and captured by oligo-dT-50 oligonucleotide probes covalently linked onto the surface of 25-channels flow-cell (step 3). A fill-in reaction is elicited with dTTP in order to fill any remaining nucleotide complementary to the poly(A) tail (step 4). Nucleic acid templates are then locked in place by the addition of dCTP, dGTP, and dATP virtual terminator (VT, here labeled B) nucleotides that inhibit extension prior to terminator cleavage (step 5). Sequencing-by-synthesis is initiated through the addition of one of the four one-color Cy-5 labeled VT nucleotide (step 6). The incorporation of fluorescence to the elongated DNA strand is measured using laser illumination and a CCD camera after unincorporated nucleotides have been rinsed. The fluorescent label is further cleaved and the incorporation of another labeled VT nucleotide is challenged. Standard sequencing runs complete 120 cycles of nucleotide additions. Ancient DNA, which is extremely fragmented, does not require further shearing before poly(A) tailing.