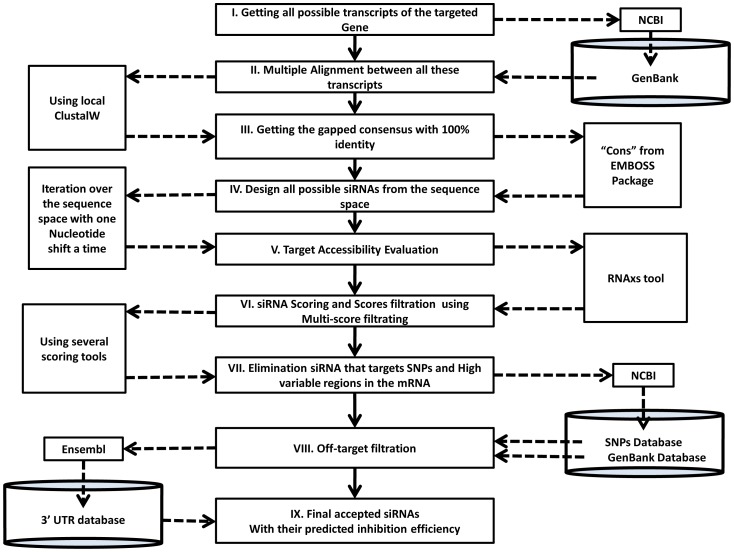
Figure 5. Flow chart of “MysiRNA-Designer” program.
MysiRNA-Designer takes an accession; get the mRNA sequence from NCBI-GenBank. Finds out if this mRNA has other transcript(s), performs multiple sequence alignment with the transcripts, if any, and takes the consensus un-gapped sequence, designs all possible siRNA in targeting sequence space available; performs target accessibility evaluation selection siRNA with energetically and structurally favored siRNA-mRNA binding. Predict siRNA efficiency using the implemented multi-score filtration; select the candidates that pass the threshold assigned for each of the ten tools used, eliminates siRNA targeting SNPs regions or off-targeted mRNA, either complete or seed homology off-target. Finally, MysiRNA-Designer shows the accepted candidates with the predicted silencing efficiency using MysiRNA-Model, it filters candidates above the assigned threshold, as the user requires.