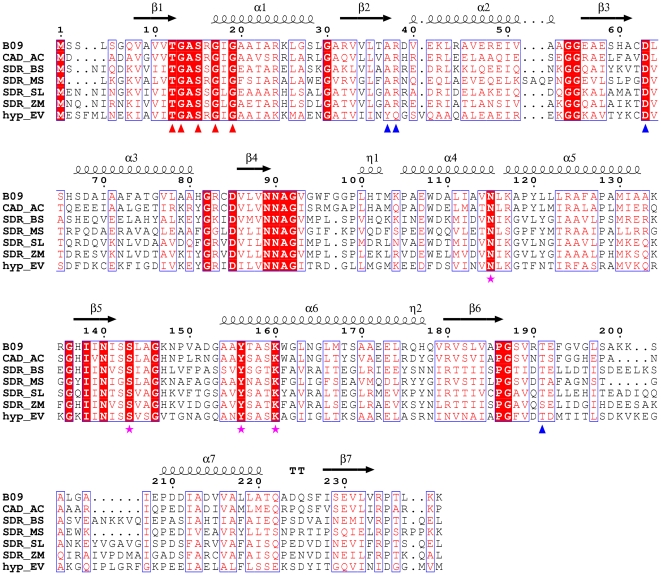
Figure 6. Structure-related functional sequence conservation between BpiB09 and homologous proteins.
Shown above the alignments are elements of the secondary structure of BpiB09. The numbering shown is from BpiB09. Red triangles indicate the dinucleotide-binding motif. Blue triangles indicate critical cofactor binding residues. Purple stars represent putative catalytic residues located at the corresponding positions of the conserved catalytic residues N115, S143, Y156, and K160, respectively, in the SDR family proteins. Strictly conserved residues are highlighted with red boxes. Biological sources and GenBank accession codes for the sequences are as follows: CAD_AC, Acidobacterium capsulatum (YP_002754051.1); SDR_BS, Bacillus subtilis subsp. Spizizenii (ZP_06875312.1); SDR_MS, Meiothermus silvanus (YP_003684578.1); SDR_SL, Spirosoma linguale (YP_003387009.1); SDR_ZM, Zymomonas mobilis subsp. Mobilis ZM4 (YP_163311.1); hyp_EV, Eubacterium ventriosum (ZP_02027436.1). Sequence alignments were assembled using T-COFFEE software (http://www.ebi.ac.uk/Tools/t-coffee) and visualized using ESPript software (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi).