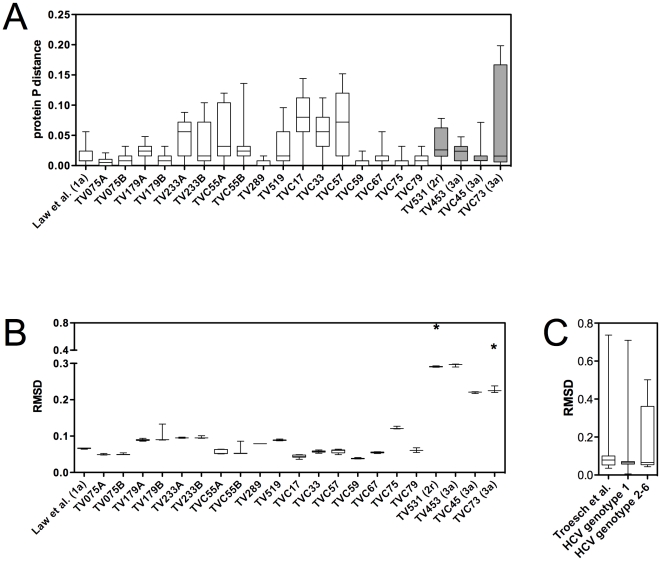
Figure 2. E2 amino-acid sequence variability and structural conservation across HCV subtypes and quasispecies.
A. Pairwise protein p distances analysis in HCV-infected subjects revealed wide disparities in E2 (residues 384–508) amino-acid sequence variability. All patients were infected with HCV-1 except TV531, who was infected with HCV-2r, and TV453, TV45, and TV73, who were infected with HCV-3a (shaded bars). B. The majority of HCV-infected subjects showed minimal structural deviation from the E2 reference structure. Asterisks (*) indicate RMSD values associated with singular outlier structures that were observed in subjects TV531 and TVC73, but not in any other subjects. C. Modelled E2 structures from HCV-infected patients from our study group (n = 391) [10] and from the HCV Database Project (genotype 1; n = 90 sequences) [36] showed less than 1 Å deviation from the reference structure. Structures based on HCV genotypes 2–6 (n = 21 sequences) [36] also showed minimal structural variation. p: protein pairwise p distance. RMSD: root-mean-square deviation. Asterisks indicate RMSD values associated with these single outlying structures