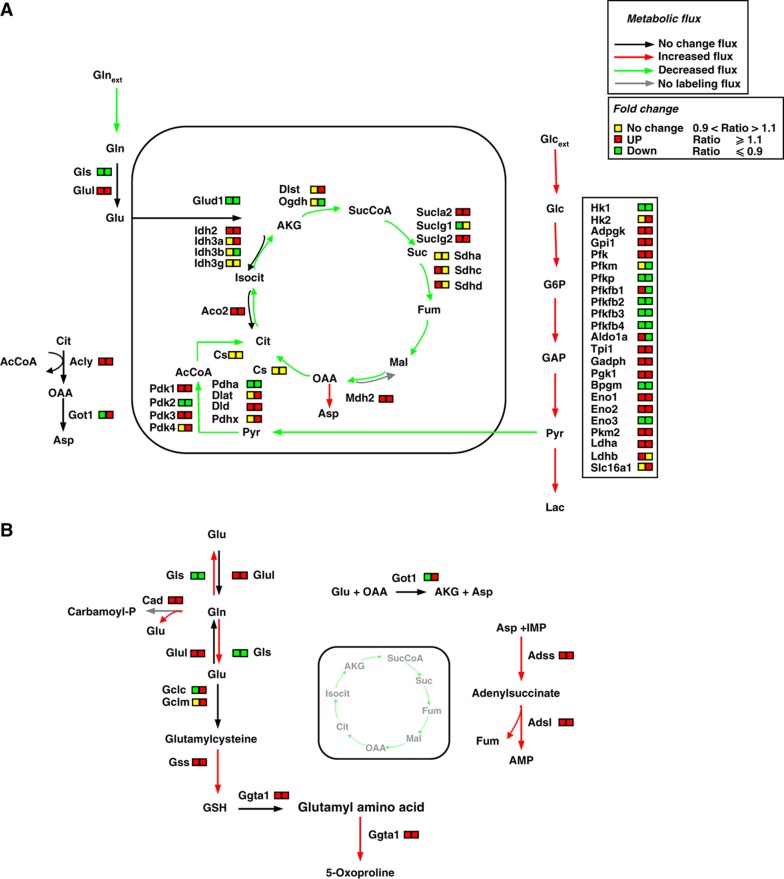
Figure 4.
Schematic integrative representation of metabolic fluxes and transcriptional data in normal and mouse K-Ras transformed cells. (A, B) Integration maps of metabolic routes identified in T cells as compared with N cells by using flux analysis and transcriptional data. The ratio between T and N cells of both metabolic fluxes and gene expression values are represented by a color code (as indicated in the upper right legends) as follow: black (no change value), red (increased value), and green (decreased value) for flux analysis and Up—red color—T/N ratio ⩾1.1, down—green color—T/N ratio ⩽0.9 and no change—yellow color—T/N ratio between 0.9 and 1.1 for transcriptional data. The list of gene abbreviations used in the figure can be found in ‘Supplementary information’. Metabolic flux analysis of central carbon metabolism was obtained by using the data derived from [U-13C5] glutamine, a mixture of [U-13C6] glucose and 1-13C] glucose (A) and [α-15N]glutamine (B) as specific isotopic tracers. The net and exchange fluxes were calculated as described under ‘Materials and methods’. Transcriptional data of N and T regulated genes were obtained as described in ‘Materials and methods’.