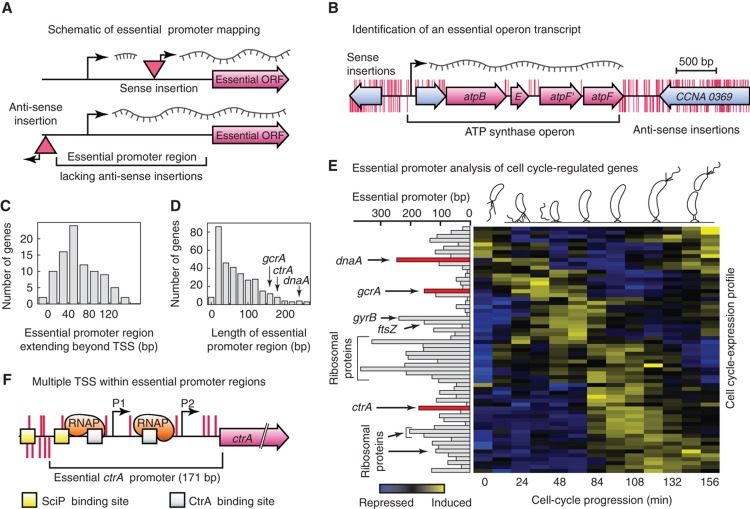
Figure 3.
Genome-wide identification of essential gene regulatory sequences. (A) Transposon insertions within the promoter region of an essential gene are only viable if the transposon-specific promoter points toward the gene (sense insertion); insertions in the opposite orientation (anti-sense insertions) are lethal. The distance between the annotated start codon and the first detected occurrence of an anti-sense insertion within the upstream region of an essential gene defines its essential promoter region. (B) Anti-sense insertions (red marks below genome track) are absent throughout the entire ATP synthase operon while sense insertions (red marks above genome track) are only tolerated within the non-essential lead gene and within non-coding regions of co-transcribed downstream genes. (C) Lengths of promoter regions that extend upstream of predicted transcriptional start sites. (D) Size distribution of essential promoter regions. The cell-cycle master regulators ctrA, dnaA and gcrA, which are subjected to complex cell cycle-dependent regulation, ranked among the longest essential promoter regions identified. (E) Essential cell cycle-regulated genes are clustered according to their temporal expression profile. Essential genes with long essential promoter regions indicate cell-cycle hub nodes subjected to complex transcriptional regulation. (F) Only insertions in the sense strand (red lines above the genome track) are tolerated within the 171 bp long essential promoter region of the cell-cycle master regulator ctrA. Insertions in the anti-sense strand (red lines below the genome track) are absent from the essential ctrA promoter. Both transcription start sites (arrows P1, P2), two DNA binding sites for CtrA (gray boxes) and one of two reported binding site for the SciP transcription factor (yellow boxes) are contained within the identified essential promoter region.