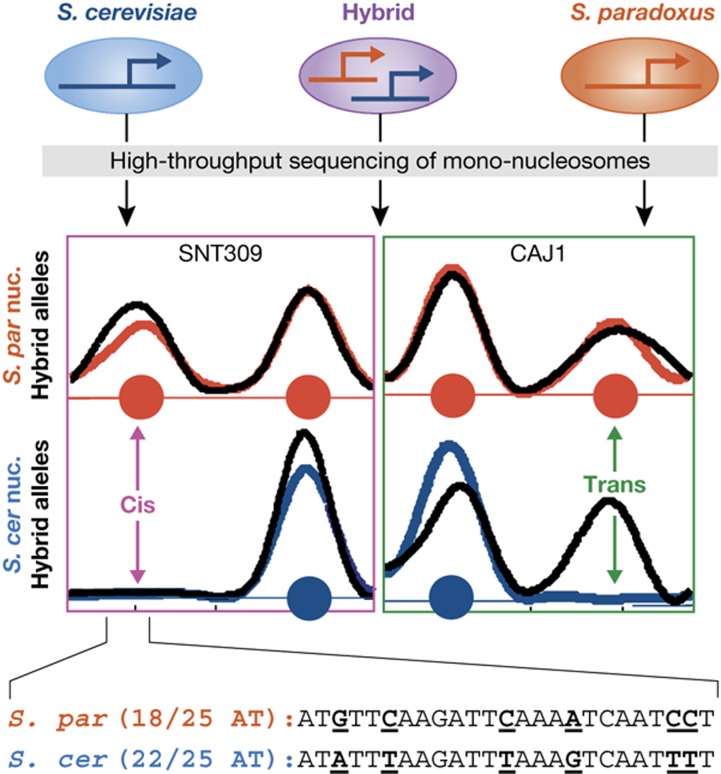
Figure 4.
Inter-species and hybrid analysis uncovers determinants of nucleosome positioning. Inter-species comparison (blue and red correspond to S. cerevisiae and S. paradoxus, respectively) and hybrid analysis (black curves) of nucleosome positioning characterizes changes due to cis and trans mutations (Tirosh et al, 2010b). Nucleosomes found in only one of the species and in the corresponding hybrid allele reflect the effect of local (cis) mutations (right). Nucleosome found in only one of the species but in both hybrid alleles reflect the trans effect of distal mutations through the activity of a chromatin-related protein or RNA (left). Sequence analysis at positions of cis changes can suggest which sequence patterns influence nucleosome positioning. For example, the inset shows sequences of the two species within a region that is bound by a nucleosome only in S. paradoxus, demonstrating that inter-species substitutions changed the frequency of AT bases between 22/25 (S. cerevisiae) and 18/25 (S. paradoxus). This is consistent with a nucleosome-disfavoring effect of AT-rich sequences, as observed systematically for inter-species nucleosome differences, and is also observed in other studies (Tillo and Hughes, 2009).