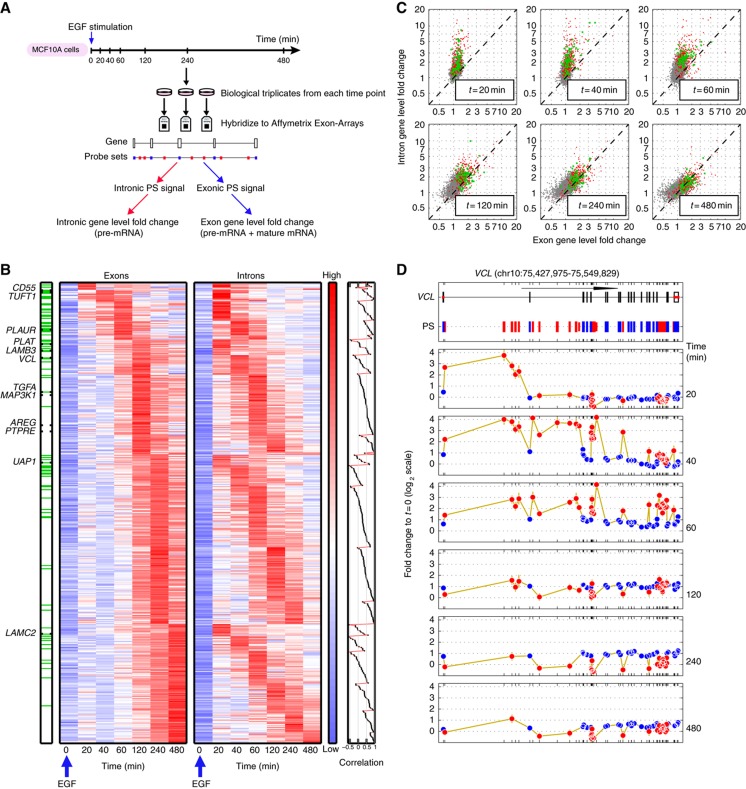
Figure 2.
EGF induces distinct transcriptional dynamics of pre-mRNAs and mRNAs in MCF10A cells. (A) Experimental outline: MCF10A mammary epithelial cells were stimulated with EGF for the indicated intervals and RNA was hybridized to Affymetrix Exon Arrays. Annotation and signal intensity-based filtering was performed to define probe sets (PS) interrogating exons (blue marks) and introns (red). Fold changes (FCs) of intronic and exonic PS were weighed separately to define the pre-mRNA and mRNA FC for each gene. (B) Pre-mRNA and mRNA profiles of 441 transcriptionally induced genes. The left (right) heatmap displays mRNA (pre-mRNA) expression, as defined by the gene-level exon (intron) FC profiles; genes are grouped according to the peak time of their mRNA expression and within each such group ordered according to the peak time of their pre-mRNA FC. Finally, within each subgroup the transcripts were ordered according to the correlation between the pre-mRNA and mRNA profiles (rightmost panel). The order of genes is the same in both heatmaps. Note that the log-transformed expression values of each row were centered and normalized (separately for pre-mRNA and mRNA, owing to their different dynamic range). Hence, the log-FC values at t=0 are not uniformly 0. Green lines on the bar on the left indicate genes with production overshoot (i.e., the maximal FC of intronic PS exceeded that of exonic PS by >2-fold). Genes chosen for detailed validation (Figure 3) are indicated. (C) Scatter plots comparing gene-level intron (pre-mRNA) and exon (mRNA) FC during EGF stimulation. Genes with mRNA FC⩽1.5 over the whole time course (grey dots), transcriptionally induced genes with production overshoot (green dots) or without (red dots) are shown. Note the similarity of mRNA FC of overshooting and non-overshooting genes. (D) Space-time description of probe-level FC of an overshooting gene. Upper panel: Genomic organization of the vinculin (VCL) gene (122 kb). Arrow length corresponds to 50 kb. Positions of exonic (blue) and intronic (red) PS are indicated. Lower panels: FC (log2 scale) of each PS with respect to its baseline value is shown for the time course outlined in (A). Only PS with present calls in all replicates of the respective time points compared are shown. Error bars (in gold) represent standard deviations. Note that intronic and exonic PS display different behavior and dynamic range.