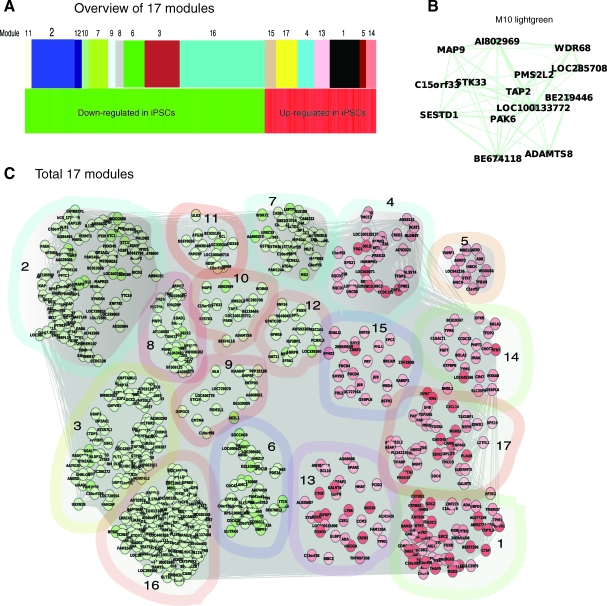
FIG. 2.
Gene networks are differentially expressed in human iPSCs and ESCs. (A) An overview of 17 network modules identified by weighted gene co-expression network analysis (Table 1 for detail). (B) An example of a module (light green) shows network component connections. Node color denotes differential expression level (iPSCs/ES), green for downregulation, and red for upregulation. Node size represents the importance of a node; bigger size indicates more importance. Edge denotes interaction strength, thicker for stronger interactions. (C) A holistic view of all 17 modules. Ten and 7 out of total 17 modules are downregulated (green nodes) and upregulated (red nodes) in iPSCs, respectively. The same illustration strategy was used in all network figures in this study. Color images available online at www.liebertonline.com/scd