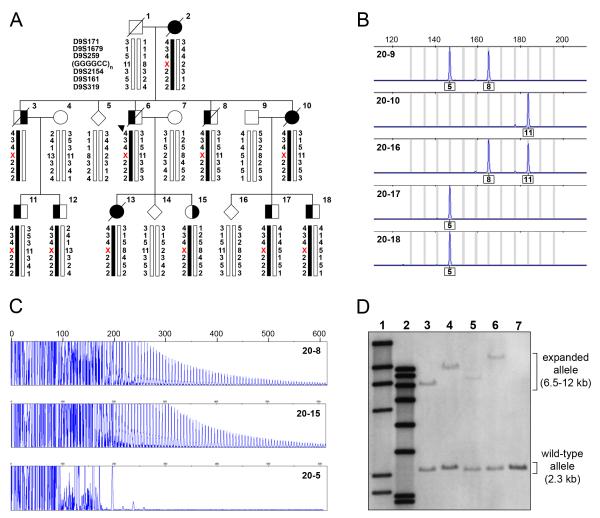
Figure 2. Expanded GGGGCC hexanucleotide repeat in C9ORF72 causes FTD and ALS linked to chromosome 9p in family VSM-20.
(A) Segregation of GGGGCC repeat in C9ORF72 and flanking genetic markers in disguised linkage pedigree of family VSM-20. The arrowhead denotes the proband. For the GGGGCC repeat, numbers indicate hexanucleotide repeat units and the X denotes that the allele could not be detected. Black symbols represent patients affected with frontotemporal dementia (left side filled), amyotrophic lateral sclerosis (right side filled) or both. White symbols represent unaffected individuals or at-risk individuals with unknown phenotype. Haplotypes for individuals 20-1, 20-2 and 20-3 are inferred from genotype data of siblings and offspring. (B) Fluorescent fragment length analyses of a PCR fragment containing the GGGGCC repeat in C9ORF72. PCR products from the unaffected father (20-9), affected mother (2-10) and their offspring (20-16, 20-17 and 20-18) are shown illustrating the lack of transmission from the affected parent to affected offspring. Numbers under the peaks indicate number of GGGGCC hexanucleotide repeats. (C) PCR products of repeat-primed PCR reactions separated on an ABI3730 DNA Analyzer and visualized by GENEMAPPER software. Electropherograms are zoomed to 2000 relative fluorescence units to show stutter amplification. Two expanded repeat carriers (20-8 and 20-15) and one non-carrier (20-5) from family VSM-20 are shown. (D) Southern blotting of four expanded repeat carriers and one non-carrier from family member of VSM-20 using genomic DNA extracted from lymphoblast cell lines. Lane 1 shows DIG-labeled DNA Molecular Weight Marker II (Roche) with fragments of 2027, 2322, 4361, 6557, 9416, 23130 bp, lane 2 shows DIG-labeled DNA Molecular Weight Marker VII (Roche) with fragments of 1882, 1953, 2799, 3639, 4899, 6106, 7427, and 8576 bp. Patients with expanded repeats (lanes 3-6) show an additional allele from 6-12kb, while a normal relative (lane 7) only shows the expected 2.3kb wild-type allele.