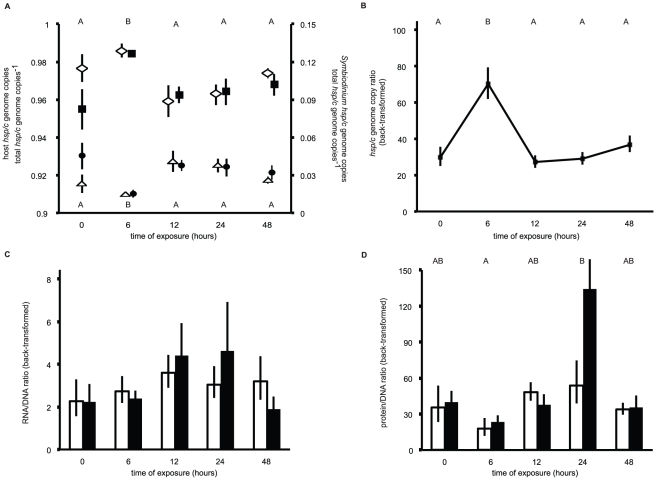
Figure 4. Genome copy proportions (GCP), genome copy ratio (GCR), RNA/DNA and protein/DNA ratios from the thermal stress experiment.
(A) Host (left y-axis) and Symbiodinium (right y-axis) hsp/c genome copies as a proportion of the total hsp/c genome copies within the sample (GCP). Hollow diamonds and triangles represent data from the control treatment for the host and Symbiodinium GCPs, respectively. Filled squares and circles represent data from the high temperature treatment for the host and Symbiodinium GCPs, respectively. Tukey's HSD letter groups (α<0.05) above and below icons correspond to the host and Symbiodinium data, respectively, and test only temporal variation, as neither significant treatment nor interaction effects were observed for these parameters. While both host and Symbiodinium GCP appear to differ between treatments at time = 0, this difference was not statistically significant (Tukey's post hoc test, p>0.05). (B) Mean host/Symbiodinium hsp/c GCRs pooled across both treatments over time. Tukey's HSD letter groups (α<0.05) are also presented. (C) RNA/DNA ratio over time for both control (white columns) and high (black columns) temperature treatments. (D) Protein/DNA ratio over time for both control (white columns) and high (black columns) temperature treatments. Tukey's HSD letter groups test only temporal variation, as neither significant treatment nor interaction effects were observed for this parameter. While the protein/DNA ratio appears to differ between treatments at time = 24, this difference was not statistically significant (Tukey's post hoc test, p>0.05). In all panels, error bars represent standard error of the mean, and for panels B–D, the back-transformed means are presented.