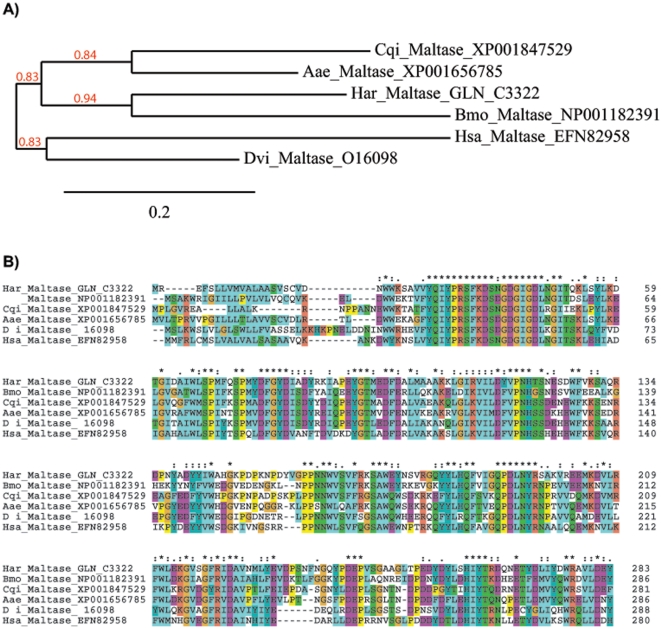
Figure 1. Phylogenetic tree and amino acid alignment of insect maltases.
(A) An unrooted Bayesian inference tree constructed from the alignment of amino acid sequences presented in (B). The Helicoverpa (Har) sequence clusters with the predicted maltase from Bombyx mori (Bmo) with good bootstrap support. (B) The complete predicted polypeptide sequences of 5 insect maltases and the identified Helicoverpa maltase are aligned. Amino acid sequence alignments were performed using MAFFT multiple alignment program. Identical residues are color-coded and residues highly conserved in all arthropod CLPs are marked with asterisks. Species abbreviations: Drosophila virilis (Dvi), Harpegnathos saltator (Hsa), Aedes aegypti (Aae), Culex quincefasciens (Cqu). GenBank accession numbers are given at the end of sequence names.