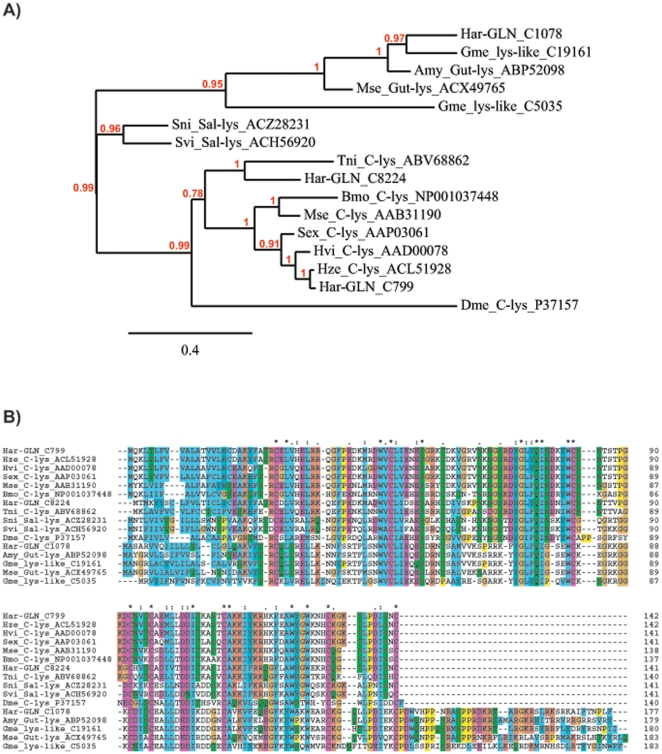
Figure 2. Gene phylogeny and amino acid alignment of C-type lysozyme protein sequences.
(A) An unrooted Bayesian inference tree constructed from the alignment of lysozyme amino acid sequences presented in (B). Bayesian posterior probabilities are shown for all major nodes supported with probability higher than 60%. (B) Amino acid alignment of the three predicted proteins from Helicoverpa (Har) together with predicted protein sequences deduced from publicly available insect sequence datasets. Amino acid sequence alignments were performed using MAFFT multiple alignment program. Identical residues are color-coded and marked with asterisks above the alignment. Species abbreviations: Galleria mellonella (Gme), Antheraea mylitta (Amy), Manduca sexta (Mse), Simulium nigrimanum (Sni), Simulium vittatum (Svi), Trichoplusia ni (Tni), Bombyx mori (Bmo), Spodoptera exigua (Sex), Heliothis virescens (Hvi), Helicoverpa zea (Hze), Drosophila melanogaster (Dme). GenBank accession numbers are given at the end of sequence names.