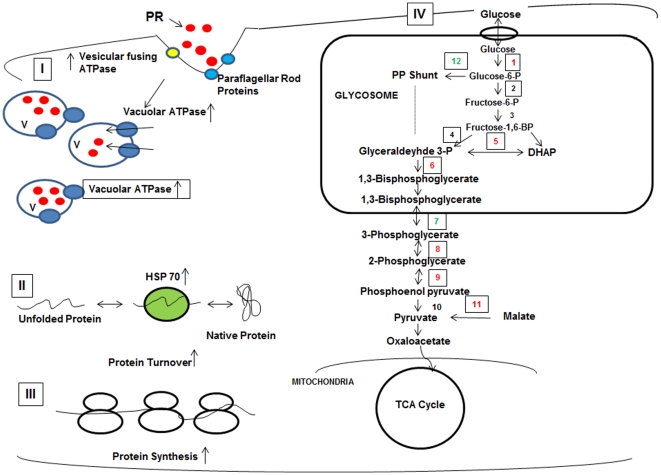
Figure 6. Proposed model for mechanism of paromomycin resistance in L. donovani.
(I) The drug paromomycin (red circles) interacts with paraflagellar rod proteins (blue circles) and prefoldin (yellow circles) and is then taken up by the Leishmania promastigotes by endocytosis. PR is then sequestered in to the vacuoles. The pump, vacuolar ATPase is found to be upregulated and the number of vacuoles is increased in the resistant parasites. (II) The chaperone proteins are found to be upregulated in the resistant cells as a result of the stress caused by paromomycin and are also involved in increased protein turnover. (III) A number of ribosomal protein subunits were found to be regulated, increasing the total protein synthesis. (IV) The glycolytic pathway was found to be upregulated. Proteins marked in red represent the upregulated proteins and the ones marked in green represent the down- regulated proteins in the case of paromomycin resistant strain. Enzymes indicated: (1) Hexokinase, (2) Phosphoglucose isomerase, (3) Phosphofructokinase, (4) Aldolase (5) Triose phosphate isomerase, (6) Glyceraldehyde 3-phosphate dehydrogenase, (7) Phosphoglycerate kinase (8) 2,3-bisphosphoglycerate independent phosphoglycerate mutase, (9) Enolase, (10) Pyruvate kinase, (11) Malic enzyme (12) glucose 6-phosphate dehydrogenase.