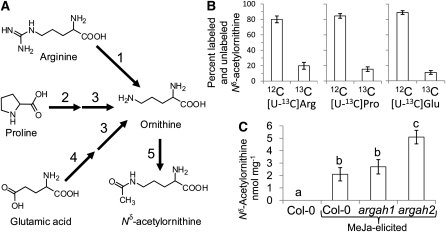
Figure 2.
Biosynthesis of Nδ-Acetylornithine from Other Amino Acids.
(A) Possible pathways leading from Arg, Pro, and Glu to Nδ-acetylornithine. 1, Arginase (At2g39020 and At4g08900); 2, Pro dehydrogenase (At3g30775 and At5g38710); 3, Orn δ-aminotransferase (At5g46180); 4, pyrroline-5-carboxylate synthase (At2g39800 and At3g55610); 5, Orn Nδ-acetyltransferase (At2g39030).
(B) Incorporation of [U-13C]Arg, [U-13C]Pro, and [U-13C]Glu into Nδ-acetylornithine in MeJA-treated Arabidopsis leaves, expressed as the percentage of labeled [13C5]Nδ-acetylornithine and unlabeled [12C]Nδ-acetylornithine in rosette leaves. Mean ± se of n = 3.
(C) Nδ-acetylornithine accumulation in arginase mutants. Mean ± se of n = 5. Letters indicate significant differences, P < 0.05, analysis of variance, followed by Tukey’s HSD test.