Abstract
RSC, Remodel the Structure of Chromatin, is an essential chromatin remodeler of Saccharomyces cerevisiae that has been shown to have DNA translocase properties. We studied the DNA binding properties of a ‘trimeric minimal RSC’ (RSCt) of the RSC chromatin remodeling complex and the effect of nucleotides on this interaction using fluorescence anisotropy. RSCt binds to 20 bp fluorescein labeled double stranded DNA with a Kd of approximately 100 nM. The affinity of RSCt for DNA is reduced in the presence of AMP-PNP and ADP in a concentration dependent manner with the addition of AMP-PNP having the more pronounced effect. These differences in the magnitude at which the binding of ADP and AMP-PNP affect the affinity of DNA binding by RSCt suggests that the physical movement of the enzyme along DNA begins between the binding of ATP and its subsequent hydrolysis. Furthermore, the fact that the highest affinity for DNA binding by RSCt occurs in the absence of bound nucleotide offers a mechanistic explanation for the low apparent processivity of DNA translocation by the enzyme.
Keywords: chromatin remodeling enzyme, RSC, DNA binding, nucleotide binding, allostery
Eukaryotic DNA is organized into a packaged structure called chromatin. The fundamental repeating unit of chromatin is a nucleosome which is made up of a histone octamer (two molecules each of the four core histones H2A, H2B, H3 and H4) and the associated DNA (1, 2). This packaging of the DNA serves two purposes: first the associated reduction in the total volume of the DNA allows it to fit within the nuclei; second this compacted structure helps to regulate the access of factors involved in transcription, replication and repair to the DNA (3, 4). Alteration of the chromatin structure is accomplished either by the covalent modification of the histones through posttranslational modifications such as acetylation, methylation and phosphorylation or through the activity of a specialized group of enzymes called chromatin remodelers (5-7). Chromatin remodelers are molecular motors that use the energy obtained from the binding and hydrolysis of ATP to alter the structure, position, and functional state the of nucleosomes (3). All chromatin remodelers share a conserved ATPase domain and are further classified into four families based on both unique domains within their ATPase subunits and unique associated subunits (3). On the protein sequence level, the ATPase domain of chromatin remodelers has been reported to have significant homology with those found in members of the families of the helicase protein (8); indeed, remodelers can be classified as members of helicase superfamily 2 (SF2). Several remodelers have been shown to be ATP-dependent DNA translocases and this DNA translocation ability forms the basis of several models for nucleosome repositioning by remodelers (3, 8-10).
RSC, Remodel the Structure of Chromatin, is an essential and abundant chromatin remodeler that has been isolated from the yeast Saccharomyces cerevisiae. RSC has been shown to reposition nucleosomes from the center of DNA fragments toward the ends without disrupting their integrity and is also capable of translocating along single- and double-stranded DNA (11-14). The ATPase subunit of RSC, Sth1, has also been shown to function as a monomeric DNA translocase like some members of the SF2 family of helicases (13). During their movement along the DNA, these translocases continually experience repeated cycles of ATP binding, ATP hydrolysis, release of ADP and inorganic phosphate, movement along the DNA, and possibly additional conformational changes (11-15). To thoroughly characterize the mechanism of ATPase-dependent DNA translocation it is therefore necessary to determine the DNA- and nucleotide-binding properties of RSC and the relation between the two; i.e., it is necessary to determine the DNA affinity of RSC in its free and various nucleotide-bound states.
In experiments reported here, a truncated version of the Saccharomyces cerevisiae RSC was purified from a bacterial expression system. Of the 15 subunits that form a complete RSC only three made up the current construct: Sth1 (301-1097 amino acids), Arp7 and Arp9. This construct is further cited as ‘trimeric minimal RSC (RSCt). The construct used in our experiments has the same RSC subunit composition as has been used by Cairns et al. to characterize the DNA translocation activity of the RSC motor (16). Sth1 is the previously mentioned ATPase subunit of RSC that also contains the DNA binding domain, and Arp7 and 9 are nuclear actin related proteins that form a stable heterodimer and have been found to be the essential components of the RSC chromatin remodeling complex and also the SWI/SNF complex (11, 15, 17). A fluorescein-labeled double-stranded DNA molecule was used as a probe to study equilibrium DNA binding by RSCt by monitoring the changes in the fluorescence anisotropy of the DNA molecule associated with the binding of RSCt to the DNA. These experiments were then repeated in the presence of varying concentrations of ADP and AMP-PNP to determine how the binding of these nucleotides by RSCt affected the DNA binding affinity of RSCt. AMP-PNP was used as it was found to mimic the ATP in ATPase activity assays as judged by its ability to inhibit the DNA-stimulated ATPase activity of RSCt more than other several screened ATP analogs. Interestingly, the binding of AMP-PNP by RSCt was associated with a significant reduction in the DNA binding affinity of RSCt. Based upon this result we propose a role for energy obtained from the nucleotide binding/ATP binding in translocation: this energy either weakens the RSCt-DNA contacts to facilitate translocation or is used as the final step of a single translocation cycle to re-establish the system for subsequent translocation.
MATERIALS AND METHODS
Protein Purification
RSCt was over-expressed and purified from a bacterial over-expression system. The CDF Duet-1 vector (Novagen) bearing a Sth1 construct (301-1097aa) with a 10X histidine tag at the N-terminus was transformed into an E. coli BL21(DE3) codon plus strain along with the RSF Duet vector containing Arp9 and Arp7 constructs. These were selected on streptomycin and kanamycin plates. The cells were grown in nutrient-rich auto-inducible media at 37°C for 4hrs, 30°C for 12 hrs and at 22°C for 24 hrs, harvested by centrifugation at 6000g at 4°C, resuspended in lysis buffer (50 mM phosphate buffer pH 7.5, 300 mM NaCl, 10% glycerol, 0.5 mM β-mercaptoethanol and 1X protease inhibitors), sonicated at 30 % duty cycle for 30 secs 10-15 times. During and after sonication, the cells were kept on ice. The lysate was then centrifuged at 20,000g for 30 minutes at 4°C. The supernatant was run on a pre-packed Ni-NTA column in an AKTA Express system and the column was equilibrated with 20 mM Tris pH 7.5, 100 mM NaCl, 10% glycerol, 0.5 mM β-mercaptoethanol, 30 mM imidazole and 1X protease inhibitors. The protein was eluted in a gradient using buffer containing 20 mM Tris pH 7.5, 100 mM NaCl, 10% glycerol, 0.5 mM β-mercaptoethanol, 500 mM imidazole and 1X protease inhibitors. The protein started eluting at approximately 50% of the gradient. The purified protein was run on 10% polyacrylamide gel containing SDS. The purified fractions were pooled, concentrated and then run through a gel filtration column equilibrated with a sizing buffer (20 mM Tris pH 7.5, 200 mM NaCl, 10 % glycerol, 0.5 mM β-mercaptoethanol and 1X protease inhibitors). The fractions that eluted corresponding to RSCt were pooled and concentrated further and then frozen in liquid nitrogen to be stored at −80°C.
Oligonucleotide substrates
HPLC-grade oligonucleotides were purchased from IDT Technologies (Coralville IA). A 20 basepair (bp) double-stranded DNA (5′-CCATGTCCATGGATACGTGG-3′ and the complementary sequence) was used as the substrate for studying the interaction of the nucleic acid with RSCt; this sequence was generated randomly and selected for its relatively high GC content. Both strands were labeled with a fluorescein label on the 5′- end. The presence of the fluorophore did not affect the ATPase activity of RSCt (data not shown) and thus unlabeled DNA were used for ATPase assays reported here. The oligonucleotides were dialyzed against Millipore distilled and deionized water to remove the salts of the storage buffer and the concentration was estimated by using the molar extinction coefficient provided on the manufacturer’s information sheet. The double-stranded DNA was prepared by mixing 20 μM of complementary strands in annealing buffer (10mM HEPES, pH 7.0 , potassium acetate 40 mM) and annealed by heating to 90°C followed by slow cooling at room temperature for several hours.
ATPase assay
ATPase activity was measured at 30°C by monitoring the hydrolysis of [α-32P] ATP. The reaction was carried out in a buffer containing 10mM HEPES pH 7.0, 1 mM ATP, 5mM MgCl2, 20mM potassium acetate, 4% glycerol, 0.5 mM DTT and 0.1mg/ml BSA. RSCt at 50 nM concentration was pre-incubated with the required concentration of double-stranded DNA and the reaction was initiated by the addition of ATP. Aliquots of the reaction samples were removed at fixed time points and titrated with an equal volume of 0.5 M EDTA was added to stop the reaction. For identifying an analog that would maximally inhibit the ATPase rate, 50 nM RSCt, 200 nM 60bp double-stranded DNA and 1 mM ATP were measured alone and in the presence of 0.5 mM potential ATPase inhibitors (AMP-PNP, ATP-γ-S and [AlF4] ADP). [AlF4] ADP was used as a mixture of equimolar concentrations of ADP and AlCl3 with a five-fold excess of NaF. The reaction products were separated by TLC on PEI Cellulose F sheets in 0.6 M potassium phosphate buffer, pH 3.4 and quantified with a PhosphorImager (GE Healthcare). ImageJ was used to quantify the intensity of the separated bands on the TLC plates. The rate of ATP hydrolysis was estimated by plotting the amount of ADP produced as a function of time using KaleidaGraph 4.0.3.0.
Fluorescence anisotropy measurements
Fluorescence anisotropy measurements were performed with a VARIAN Cary Eclipse fluorescence spectrometer. The excitation and emission wavelengths for the fluorescein fluorophore were 485 nm and 518 nm, respectively, and a 5 nm slit width was used for both excitation and emission. The measurement was started two minutes after the addition of the titrant (protein or nucleotide). Triplicate measurements of the sample were automatically recorded by instrument and the average of these three measurements was taken for each anisotropy value.
The titration experiments were conducted at 30°C using a 3 ml cuvette with the same buffer used for the ATPase activity. Protein was added into a 2.8 ml solution of 50 nM 20bp fluorescein-labeled DNA in a stepwise manner up to 12X the total DNA concentration. The experiments in the presence of nucleotide were conducted similarly with the exception that the nucleotide at a specified concentration was added into the DNA solution prior to the protein titration. The effect of the nucleotide on the preformed protein-DNA complex was measured through the titration of increasing amounts of nucleotide into solution containing a pre-formed RSCt-DNA complex as the substrate for estimation of anisotropy changes; this complex was formed by pre-incubating 50 nM 20 bp DNA and 500 nM RSCt which had been pre-incubated together. The change in concentrations due to dilution during the titration was taken into account during data analysis.
Data analysis fluorescence anisotropy measurements
Although the contact size for RSC binding to double-stranded DNA has been estimated to be (8 ± 3) bp (18) there has been no estimate of the occluded site size RSC. Because of this we decided to measure the equilibrium binding of RSCt to a 20bp DNA molecule in order to minimize the potential of multiple RSCt complexes binding to the same DNA molecule. We nevertheless analyzed the equilibrium binding isotherm for the RSCt-DNA interaction to models that included the possibility of multiple RSCt complexes being bound to the same DNA molecule. The best global fit of all binding isotherms conducted at different total concentrations of DNA and RSCt corresponded to a 1:1 binding model, with an decreasing quality of fit as judged by the variance, with each additional possible binding site; for example, the variance of the fit for a 1:2 RSCt:DNA binding model was a factor of 10 larger than the variance of the fit for 1:1 RSCt:DNA binding model. Similar results were obtained regardless of the level of positive or negative cooperativity that were included in the fit. Furthermore, despite the lack of evidence for self-association of the RSCt trimer, we also tested binding models which included oligomerization of RSCt free in solution and DNA-binding-stimulated oligomerization of RSCt. Again, the best fit corresponded to a model which did not include any self-association of RSCt. Because of these results we are confident that RSCt binds to the 20 bp DNA molecule as a 1:1 interaction and subsequently used that model to analyze our data.
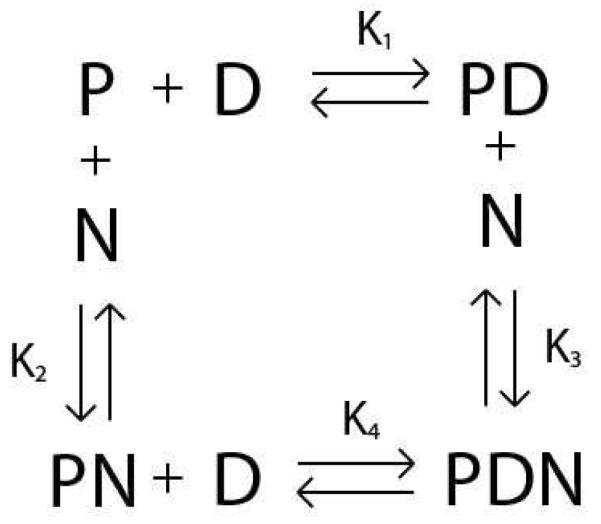
The various interactions involved in this model are shown in Scheme 1. As shown in Scheme 1: K1 is the equilibrium association constant for the RSCt-DNA interaction when no other co-factor/ligand/nucleotide is present, K2 is the equilibrium association constant for the RSCt-nucleotide interaction, K3 is the equilibrium association constant for the preformed RSCt-DNA complex interaction with the nucleotide, and K4 is the equilibrium association constant for the RSCt and DNA interaction in presence of nucleotide.
Scheme 1.
The value of the anisotropy measured for the free DNA at the start of a titration was subtracted from all subsequent anisotropy values measured for that titration. The subsequently determined change in anisotropy was therefore a function of only the concentrations of the PD and PDN species (see Scheme 1).
| (1) |
In equation (1), f is the change in the anisotropy of the sample (from the anisotropy measured for DNA alone), [PD] and [PDN] are the concentrations of the PD and PDN complexes, respectively (see Scheme 1). [Dtotal] is the total DNA concentration, s is the anisotropy associated with the PD complex and A is the ratio of the change in anisotropy associated with the PDN complex (compared with the anisotropy of the free DNA) to that associated with the PD complex; i.e., a value of A = 1 would mean that PD and PDN have the same change in anisotropy. The factor A is included to allow for the possibility that the binding of the nucleotide affects the structure of the protein-DNA complex; for example, the binding of the nucleotide might alter the structure of RSCt and thereby change its rotational diffusion rate.
Using Scheme 1 we can rewrite equation (1) in terms of the equilibrium association constants.
| (2) |
In equation (2), K1, K2, and K4 are the previously described equilibrium association constants and [P], [D], and [N] are the concentrations of free protein, free DNA, and free nucleotide, respectively.
Estimates of these equilibrium association constants and species concentrations were obtained through simultaneous global implicit analysis of the binding isotherms using the following equations:
| (3) |
| (4) |
| (5) |
In equations (3) through (5), [Ptotal] and [Ntotal] are the total protein and total DNA concentrations, respectively. An estimate of K3 can be obtained using the thermodynamic constraint that ΔG° = 0 for a closed pathway.
| (6) |
All data analysis was performed using Conlin, kindly provided by Dr. Jeremy Williams (19). All reported uncertainties in this manuscript represent 68% confidence limits (±1 standard deviation) and were determined by performing a 100 cycle Monte Carlo (20) simulation built into Conlin.
RESULTS
RSC is an ATP dependent translocase that uses energy obtained from ATP binding and hydrolysis to move along DNA. In order to more fully understand this energy transduction process and the associated DNA translocation mechanism, we must determine how the affinity of RSC for the DNA changes during the ATPase cycle. Although an estimate of the DNA binding affinity of RSC has previously been reported (15), the allosteric regulation of this affinity by nucleotide binding is unknown.
DNA binding properties of RSCt
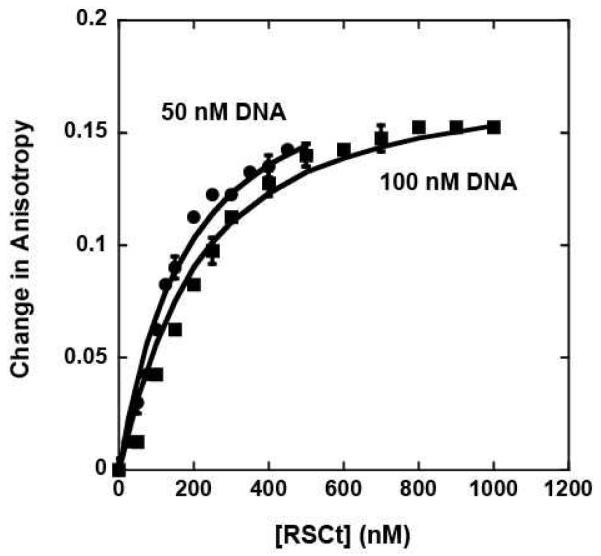
All of our DNA binding experiments were conducted using a 20 bp DNA molecule (see Material and Methods) in which one or both strands were labeled on the 5′ end with a fluorophore. The binding of RSCt to this DNA molecule was monitored by changes in the fluorescence anisotropy of the fluorophore that occur upon RSCt binding; no significant change in the fluorescence intensity of the fluorophore was observed upon RSCt binding. As shown in Figure 1, the binding of RSCt to the DNA resulted in an increase in the anisotropy of the fluorophore, consistent with the formation of the complex. In order to determine the model for the RSCt-DNA interaction we performed simultaneous global analysis of equilibrium binding isotherms collected at different total DNA concentrations (see Figure 1). As discussed in Materials and Methods the simplest model consistent with this analysis of the data is a 1:1 binding model (see Scheme 1) and thus this model was used for all subsequent analysis of our data. The average value of the five independent estimates of K1 that we obtained from our analysis of the data in Figures 1, 3, and 4 using Scheme 1 (equations (2) through (6)) is K1 = (7 ± 1) × 106 M−1, corresponding to an equilibrium dissociation constant of Kd = (140 ± 20) nM. The solid lines in Figure 1 are the corresponding fits. In a separate set of experiments, we included a 20 bp DNA without a fluorophore label as a competitor molecule. Through global analysis of the binding of RSCt to the fluorophore labeled DNA as a function of the concentration of unlabeled DNA in the solution we were able to determine an equilibrium association constant of K1 = (3.49 ± 0.13) × 106 M−1 for RSCt binding to unlabeled DNA (data not shown). Thus, the presence of the fluorophore increases the affinity of the RSCt for binding to the DNA, consistent with what has been observed in previous anisotropy based measurements of DNA binding by helicases (21).
Figure 1.
Fluorescence anisotropy measurements of RSCt binding to DNA in the absence of nucleotide. The binding experiment involved monitoring the changes in the anisotropy of the fluorescein labeled 20 bp DNA as a function of both RSCt and DNA concentration as described in Materials and Methods. Binding isotherms collected with initial DNA concentrations of 50 nM and 100 nM were analyzed simultaneously using equations (2) though (6) to determine an estimate of (140 ± 20) nM for the Kd of the interaction. The solid lines are the associated fits of this data to this model.
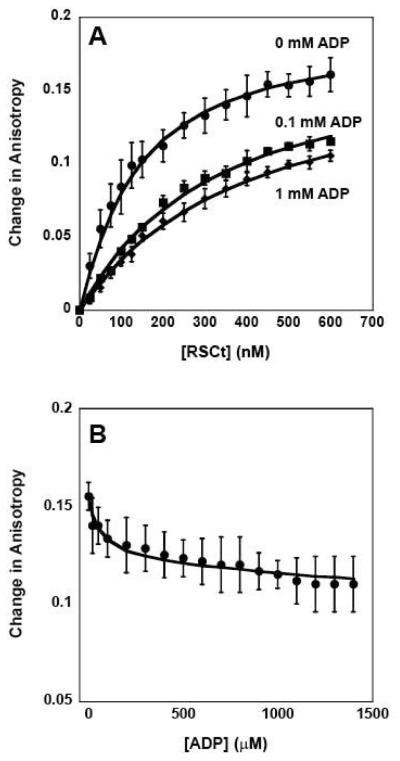
Figure 3.
Fluorescence anisotropy measurements of the equilibrium DNA binding by RSCt in presence of ADP. (A) Representative traces of binding isotherms collected in the presence of 0 mM ADP (circles), 0.1 mM ADP (squares), and 1 mM ADP (diamonds). In these experiments the total initial DNA concentration was 50 nM. (B) Representative trace of binding isotherms collected in experiments in which pre-formed RSCt-DNA complexes were titrated with ADP. In these experiments the intial RSCt and DNA concentrations are 500 nM and 50 nM, respectively. The solid lines in both panels are the associated fits of all data to Scheme 1 (equations (2) through (6)).
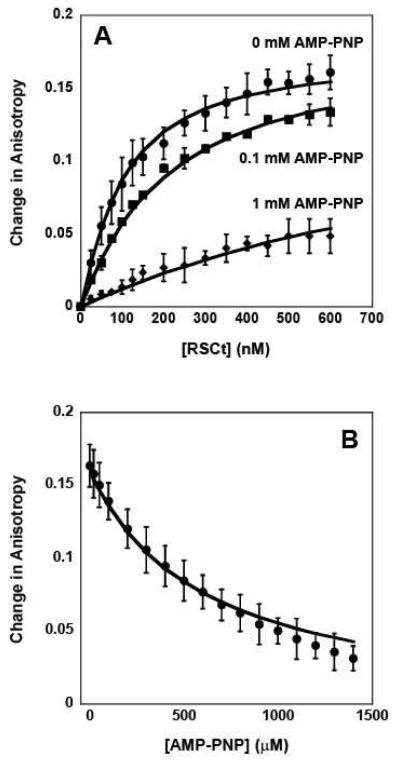
Figure 4.
Fluorescence anisotropy measurements of the equilibrium DNA binding by RSCt in presence of AMP-PNP. (A) Representative traces of binding isotherms collected in the presence of 0 mM AMP-PNP (circles), 0.1 mM AMP-PNP (squares), and 1 mM AMP-PNP (diamonds). In these experiments the total initial DNA concentration was 50 nM. (B) Representative trace of binding isotherms collected in experiments in which pre-formed RSCt-DNA complexes were titrated with AMP-PNP. In these experiments the intial RSCt and DNA concentrations are 500 nM and 50 nM, respectively. The solid lines in both panels are the associated fits of all data to Scheme 1 (equations (2) through (6)).
Screening for an ATP analog
RSCt is a DNA stimulated ATPase that uses the energy obtained from the binding and hydrolysis of ATP to translocate along the DNA molecule. Because of this, it is not possible to determine equilibrium constants for DNA binding by RSCt in presence of ATP. Thus, in order to characterize equilibrium binding in the presence of ATP, we decided to measure equilibrium DNA binding by RSCt in the presence of slowly hydrolyzable ATP analogs that would bind to RSC in a manner similar to ATP. In order to determine which ATP analog would most effectively mimic ATP for binding to RSCt, we screened a series of ATP analogs for their ability to inhibit the DNA stimulated ATPase activity of RSCt (see Materials and Methods). The results of this screen (Figure 2) indicate that AMP-PNP is the most effective inhibitor of the DNA-stimulated ATPase activity of RSCt and thus effectively mimics ATP when binding to RSCt. Interestingly, although ATP-γ-S has been used by Lorch et al (12) in their study we found AMP-PNP to be more efficient inhibitor as compared to ATP-γ-S. Although [AlF4]-ADP appears to be the most efficient inhibitor but as we could see comparable inhibition in presence of AlCl3 also, which is a component of the complex. Thus, the inhibition observed in the presence of AlCl3 may result from a general ionic-based destabilization of the RSCt-DNA complex or the RSCt-ATP binding itself, rather than direct competition with ATP for the nucleotide binding site of RSCt. Therefore, we chose to work with AMP-PNP as an ATP analog. Not surprisingly, ADP was also found to be an efficient inhibitor of the ATPase activity.
Figure 2.
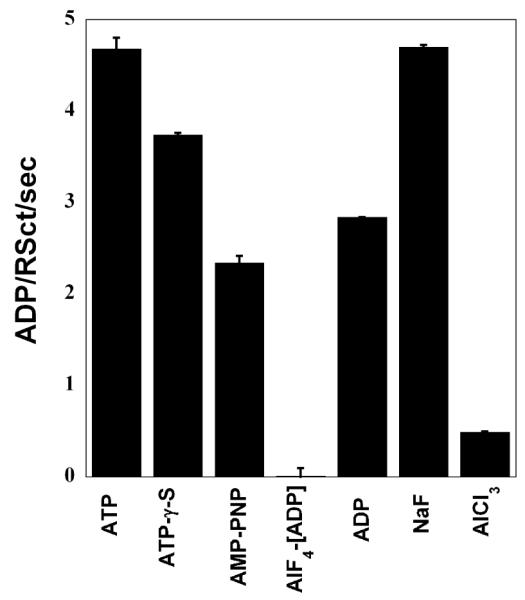
Effect of ATP analogs on ATPase activity of RSCt. The ATPase rate of 50 nM RSCt, 200 nM 60 bp ds DNA and 1 mM ATP was measured alone, in presence of 0.5 mM potential ATPase inhibitors. [AlF4] ADP was used as a mixture of equimolar concentrations of ADP and AlCl3 with a five-fold excess of NaF. AMP-PNP was found to be the best mimic of ATP for binding to RSCt. The results presented are the average values of three experiments.
DNA binding in presence of ADP and ATP analog
In order to determine the effects of nucleotide binding by RSCt on its affinity for binding to DNA we conducted a series of experiments in which the RSCt-DNA binding interaction was measured as a function of added nucleotides. In the first set of experiments, ADP concentrations up to 1.6 mM were included in the reaction. As shown in Figure 3A, the affinity of RSCt for the DNA is reduced in the presence of ADP. To further evaluate the effect of ADP binding on the RSCt-DNA interaction we performed a separate set of experiments in which we monitored the dissociation of a pre-formed RSCt-DNA complex resulting from the addition of ADP. In these experiments, the complex was titrated with ADP and the resulting changes in the fluorescence anisotropy of the DNA were monitored. The results of these experiments, shown in Figure 3B, show that the addition of nucleotide resulted in a decrease in anisotropy, which would correspond to a decrease in the fraction of DNA bound by RSCt. Thus, consistent with the results of our previous experiments, these results suggest that the affinity of DNA binding by RSCt is reduced when ADP is bound. The data in figure 3, together with six additional RSCt-DNA binding isotherms collected at varying concentrations of ADP ranging from 0.1 mM to 1 mM (data not shown), were simultaneously analyzed according to Scheme 1 (equations (2) through (6)) using non-linear least squares (NLLS) to determine estimates of the equilibrium association constants. The results of our initial analysis of the data, summarized in Tables 1 and 3, returned an estimate of K1 = (10.6 ± 0.6) × 106 M−1 for the affinity of the RSCt-DNA interaction. This estimate of K1 is higher than, but still consistent with, the value that we obtained in the analysis of the binding isotherms from experiments in which the total DNA concentration was varied. This suggests that the same 1:1 binding model is consistent with binding isotherms collected in the presence of ADP; i.e., the presence of the nucleotide has not altered the stoichiometry of the binding interaction.
Table 1.
Results of global NLLS analysis of 9 independent RSCt-DNA binding isotherms measured in the presence of varying concentrations of ADP in the range from 0.1 mM to 1.0 mM using equations (2) through (6). Column A corresponds to global fits in which the variable A (see materials and methods) was allowed to float and column B corresponds to global fits in which the variable A was fixed to unity.
| A | B | |
|---|---|---|
| K1 (M−1) | (10.6 ± 0.6) × 106 | (9.0 ± 0.6) × 106 |
| K2 (M−1) | (5.0 ± 1.2) × 104 | (7 ± 2) × 104 |
| K3 (M−1) | (9 ± 3) × 103 | (2.2 ± 0.6) × 104 |
| K4 (M−1) | (2.01 ± 0.13) × 106 | (2.7 ± 0.1) × 106 |
| A | 1.28 ± 0.05 | 1.0 (fixed) |
| s | 0.160 ± 0.013 | 0.177 ± 0.013 |
| Variance | 8.5 × 10−6 | 1.2 × 10−5 |
Table 3.
| ADP | AMP-PNP | ADP | AMP-PNP | |
|---|---|---|---|---|
| 1/K1 | (99 ± 5) nM | (95 ± 3) nM | ||
| 1/K2 | (14 ± 4) μM | (72 ± 6) μM | (20 ± 5) μM | (130 ± 12) μM |
| 1/K3 | (45 ± 12) μM | ≥ 2.4 M | (110 ± 40) μM | (920 ± 120) μM |
| 1/K4 | (370 ± 14) nM | ≥ 2.5 mM | (500 ± 30) μM | (680 ± 90) μM |
| A | 1.0 (fixed) | 1.28 ± 0.05 | 0.00 ± 0.03 | |
As shown in Table 1, the results of this global analysis also indicate that RSCt is capable of binding ADP with reasonable affinity, K2 = (5.0 ± 1.2) × 104 M−1, and that this affinity is reduced by approximately five fold when RSCt is bound to DNA, K3 = (9 ± 3) × 103 M−1. Consequently, the affinity of RSCt for binding DNA is reduced by approximately a factor of five when RSCt is already bound to ADP, K4 = (2.01 ± 0.13) × 104 M−1. The results of this global analysis also estimated a value of A = (1.28 ± 0.05), suggesting that the binding of nucleotide by RSCt might change the structure and associated rotational diffusion rate of the complex. Specifically, this estimate of A would be consistent with RSCt being less compact after binding ADP. It is important to note, however, that the estimates of A and K4 obtained from an analysis of binding isotherms using equations (2) through (5) are highly correlated with each other; the correlation coefficient was determined to be −0.93. Furthermore, the correlation coefficient between K2 and K4 was determined to be 0.22. Thus, independent estimates of these parameters are subject to some error owing to their mutual correlation.
In order to further explore the robustness of these parameter estimates we reanalyzed our data with the value of A constrained to be unity. This analysis resulted in a fit of only slightly poorer quality, as judged by its variance, but with similar estimates of the equilibrium association constants (see Tables 1 and 3); the solid lines in Figure 3 are the fits obtained in this second analysis. Regardless of the possibility of correlation between the estimates of A and the equilibrium binding constants, however, the conclusion from the analysis is the same: in the presence of ADP the affinity of RSCt for binding to DNA is reduced.
We then repeated these same experiments in the presence of AMP-PNP rather than ADP. As shown in Figure 4A and 4B, the affinity of RSCt for the DNA is also reduced in the presence of AMP-PNP. The data in Figure 4, together with five additional RSCt-DNA binding isotherms collected at 0.2, 0.3, 0.4, 0.6, and 0.8 mM AMP-PNP (data not shown), were simultaneously analyzed according to Scheme 1 (equations (2) through (6)) using NLLS to determine estimates of the equilibrium association constants. The results of our initial analysis of this data, summarized in Tables 2 and 3, returned an estimate of K1 = (10.4 ± 0.5) × 106 M−1 for the affinity of the RSCt-DNA interaction. This estimate of K1 is higher than, but still consistent with, the value that we obtained in the analysis of the binding isotherms from experiments in which the total DNA concentration was varied and is within error of the estimated value obtained from the analysis of the binding isotherms collected in the presence of ADP. This suggests that the same 1:1 binding model is consistent with binding isotherms collected in the presence of AMP-PNP; i.e., the presence of the nucleotide has not altered the stoichiometry of the binding interaction.
Table 2.
Results of global NLLS analysis of 9 independent RSCt-DNA binding isotherms measured in the presence of varying concentrations of AMP-PNP in the range from 0.1 mM to 1.0 mM using equations (2) through (6). Column A corresponds to global fits in which the variable A (see materials and methods) was allowed to float and column B corresponds to global fits in which the variable A was fixed to unity.
| A | B | |
|---|---|---|
| K1 (M−1) | (10.4 ± 0.5) × 106 | (13 ± 1) × 106 |
| K2 (M−1) | (7.7 ± 0.7) × 103 | (1.39 ± 0.12) × 104 |
| K3 (M−1) | (1.09 ± 0.14) × 103 | ≤ (4.22 ± 0.18) × 10−1 |
| K4 (M−1) | (1.47 ± 0.19) × 106 | ≤ 4 × 102 (fixed) |
| A | 0.00 ± 0.03 | 1.0 (fixed) |
| s | 0.186 ± 0.015 | 0.173 ± 0.014 |
| Variance | 8.0 × 10−6 | 1.75 × 10−5 |
As shown in Table 2, the results of this global analysis also indicate that the affinity of RSCt for binding AMP-PNP, K2 = (7.7 ± 0.7) × 103 M−1, is approximately one order of magnitude less than the affinity for binding ADP; similarly, the affinity for DNA-bound RSCt to bind AMP-PMP, K3 = (1.09 ± 0.14) × 103 M−1, is less than the affinity for DNA-bound RSCt to bind ADP. Interestingly, the results of this analysis returned an estimate of K4 = (1.47 ± 0.19) × 106 M−1, corresponding to an approximately five fold reduction of the affinity for DNA binding in the presence of AMP-PNP, and A = (0.00 ± 0.03), indicating that there is no change in anisotropy associated with the PDN species (as compared with the free DNA).
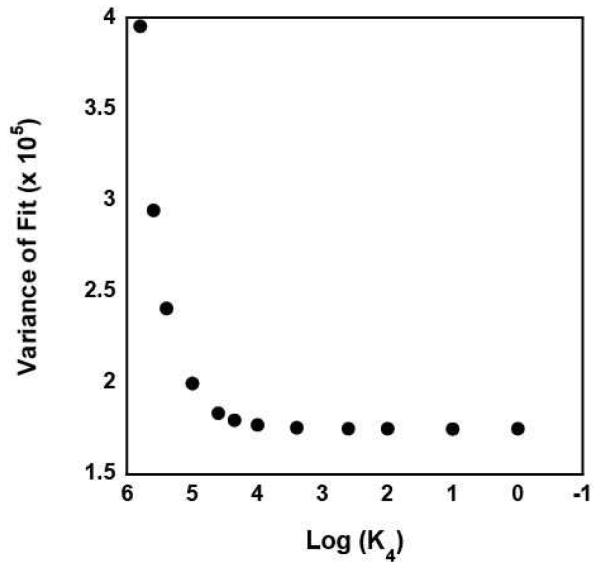
Since it is unlikely that the PDN species would have an anisotropy similar to that of free DNA the estimate of A = 0 returned from the analysis the data likely suggests that the concentration of the PDN species in the solution was too small to significantly affect the fitting. To further explore this possibility, we reanalyzed the data with A constrained to unity. The results of this analysis, however, provided no constrained estimates of K4 or K3, consistent with the previous conclusion that the concentration of the PDN species in the solution was too small to significantly affect the fitting. We subsequently performed a series of analyses of the data in which the value of K4 was constrained to different values. As shown in Figure 5, the quality of the fit of the data, as judged by its variance, improves as K4 is decreased, but becomes invariant of the value of K4 for K4 < 400 M−1 ; the solid lines in Figure 4 are the fits obtained in an analysis with K4 constrained to be 400 M−1. Thus, our analysis can provide only a limit on the value of K4 (and K3) for RSCt-DNA binding interactions conducted in the presence of AMP-PNP. This is consistent with there being only a small fraction of the RSCt-DNA-AMP-PNP complex in the solution at equilibrium.
Figure 5.
The variance of the fit of nine independent binding isotherms collected in the presence of varying concentrations of AMP-PNP as a function of the value of K4 constrained in the analysis of the data to Scheme 1 (equations (2) through (6)).
Taken together, all of these results indicate that RSCt interacts with DNA tightly in the absence of nucleotide and there is a decrease in this affinity in presence of ADP and AMP-PNP, with the effect of ATP analog being at least 1000 fold greater than that of ADP.
DISCUSSION
Several models have been proposed for the nucleosome repositioning activity of RSC and a common feature of these models is the translocation of RSC along the nucleosomal DNA (13). The translocation of RSC results in the formation of loops or bulges in the DNA over the surface of the histone octamer, the resolution of which results in the displacement of the octamer relative to the DNA and thus the repositioning of the nucleosome (12). Because DNA translocation is an ATP-dependent process, it involves repeating cycles of ATP binding, ATP hydrolysis, and product release (22). Therefore, in order to understand how the ATPase activity and the DNA translocation activity are interrelated, it is necessary to understand the role of nucleotide binding/hydrolysis in the modulation of the DNA binding affinity of RSC.
DNA binding property of RSCt
The DNA dependent ATPase activity and associated DNA translocation activity, of the full RSC complex and of the ATPase subunit of this complex, Sth1, have been published (13), and estimates of the DNA binding affinity of RSC have also been reported (23). However, a quantitative characterization of the binding affinity of RSC with DNA is still lacking. Here we detail the results of our studies of equilibrium DNA binding by a truncated version of the RSC complex, consisting of the three subunits out of a total of 15 subunits of RSC complex, and the changes in this binding interaction which occurs when nucleotides are present in the solution. The DNA-stimulated activity of RSCt has previously been found to be identical to that of RSC (16), but the higher yield of RSCt compared to RSC makes RSCt a preferred substrate for studies of DNA binding and translocation. Through implicit fitting of the resulting binding isotherms using NLLS analysis we were able to determine quantitative estimates of the equilibrium binding constants associated with these reactions (Scheme 1).
As shown in Scheme 1, four equilibrium binding constants are required to quantify the four different interaction modules between RSCt, DNA and nucleotides. From the analyses of our data we obtain five independent estimates of the equilibrium association constant for the RSCt binding to DNA: K1 = (7±1) × 106 M−1, K1 = (10.6 ± 0.6) × 106 M−1, K1 = (9.0 ± 0.6) × 106 M−1, K1 = (10.4 ± 0.5) × 106 M−1, and K1 = (13 ± 1) × 106 M−1. The weighted average of these estimates is K1 = (1.00 ± 0.03) × 107 M−1, corresponding to Kd = (100 ± 3) nM, which is comparable to what has been observed for the DNA binding affinity of other DNA translocases. Dissociation constants of 276nM, 220nM, 160nM and 2-4 nM have been reported for E. coli UvrD, E. coli Rep, Bacillus stearothermophilus PcrA and Hepatitis C NS3 helicases (22, 24-26). Lorch et al have estimated an affinity of about 10−8 M−1 or 10 nM for the RSC-nucleosome interaction from their gel mobility based experiments (23). Although these values were measured using different lengths and sequences of DNA, as well as different solution conditions, they nevertheless provide a range for the affinity of DNA translocases for DNA binding.
Based on the DNA stimulated ATPase activity, Cairns et al (13) have reported a Michaelis-Menten constant (Km) of 1.04 ± 0.13 μM for RSC with linear 45-mer DNA with the further observation that Km values were independent of the nature of the substrate. And for Sth1 with linear 45-mer DNA they have reported a Km value of (0.82 ± 0.05) μM. The differences between the Km values reported by Cairns et al and the Kd value that we estimate is consistent with our observation that the affinity of RSCt for DNA binding is decreased in the presence of ADP and AMP-PNP. Of course, a more quantitative comparison between Km and Kd are complicated by the fact that Km is a kinetic variable, associated with the steady-state translocation activity of the enzyme, whereas Kd is an equilibrium variable. Indeed, Km is a function of many processes like DNA binding, DNA dissociation, ATP binding, ATP hydrolysis etc. However, it is worth noting that in a recently published study of the DNA translocation activity of RSCt by Cairns et al. a value of KM of 0.03 mM for the dependence of the translocation rate on ATP concentration was reported (16). This value is comparable with our estimates of 1/K2 = (14 ± 4) μM and (72 ± 6) μM for RSCt binding to ADP and AMP-PMP, respectively.
The affinity of RSCt for DNA is reduced in the presence of nucleotides
The effect of nucleotide binding on the DNA binding properties of RSCt was studied by conducting DNA binding experiments in presence of AMP-PNP and ADP. Among the tested analogs AMP-PNP was found to be the best inhibitor of ATPase activity and presumably mimics the structure features of ATP in the ATP binding site of Sth1. An overall decrease in the affinity of RSCt for DNA could be observed in presence of both ADP and AMP-PNP (Figures 3 and 4); the effect was similar whether DNA was added into RSCt-nucleotide complex or the nucleotide was added into a preformed RSCt-DNA complex. However, the decrease in affinity in the presence of AMP-PNP affinity is approximately 100 times larger than the decrease in affinity in the presence of ADP.
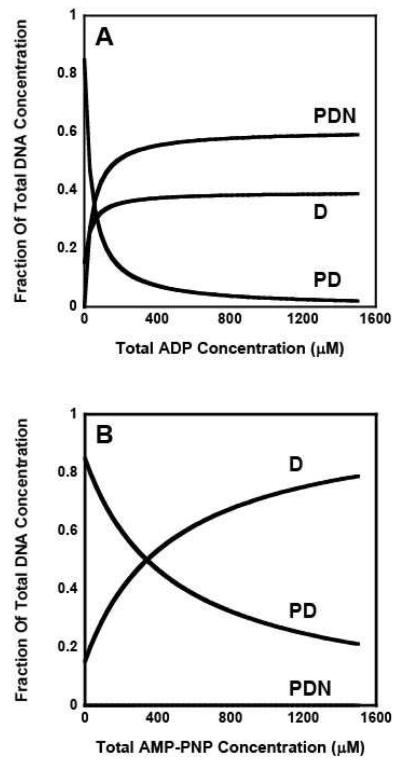
We also estimated the concentration of DNA bound to RSCt at different nucleotide concentrations during the course of these experiments using a numeric simulation with the estimated values of the equilibrium association constants. It is clearly visible that in presence of ADP (Figure 6A) a significant portion of RSCt is present as the DNA bound species whereas in the presence of AMP-PNP (Figure 6B) most of the protein is free from DNA at higher AMP-PNP concentrations. It can be argued that the binding of nucleotides to RSCt results in a conformational change and this change in conformation has an effect on the DNA binding properties. Furthermore, the extent of the conformational change produced by binding of AMP-PNP is greater as compared to one produced by ADP and this probably is a consequence of the difference in the binding of these two nucleotides.
Figure 6.
Computer simulations of the DNA bound RSCt species fractions present at different total nucleotide concentrations. A mathematical estimation of DNA bound to the RSCt as a function of nucleotide concentration was performed by using the Monte Carlo simulation using the experimental data as the input; Panel (A) is for ADP and Panel (B) is for AMP-PNP. It is clearly visible that in presence of ADP a significant portion of RSCt is present as the DNA bound species while as in presence of AMP-PNP most of the protein is unbound at higher concentrations.
It may seem counterintuitive that RSCt has DNA-stimulated ATPase activity and yet the binding of ATP by RSCt stimulates the dissociation of RSCt from DNA. There is of course no contradiction in these results provided that the rate of ATP hydrolysis by RSCt is faster than the rate of dissociation of the RSCt-ATP complex from the DNA. Indeed, although the equilibrium population of the PDN species may be very small, it can nevertheless be transiently populated to an extent and duration significant enough to give rise to the observed DNA-stimulated ATPase activity of the RSCt. In the absence of direct experimental measurements of the rates of hydrolysis and product release by RSCt during DNA translocation we can only speculate about the associated ATPase reaction cycle. However, the fact that the reported value of KM = 30 μM for the dependence of the DNA translocation rate of RSCt on ATP concentration (16) is closer to our estimate of 1/K2 = (14 ± 4) μM for RSCt binding to ADP than to our estimate of 1/K2 = (72 ± 6) μM for RSCt binding to AMP-PMP would be consistent with the ADP-bound (or ADP-Pi-bound) state being populated for a longer period of time than the ATP-bound state during the ATPase reaction cycle.
Comparison to other helicases
Chromatin remodelers are classified to be members of the SF2 superfamily of helicases based upon amino acid sequence homology, although they lack helicase activity (3, 27, 28). A common feature of several helicases is that their affinity for single- or double-stranded DNA is modulated by ATP or ADP binding (26, 29, 30). Furthermore, the allosteric regulation of DNA binding by nucleotide binding has formed the basis for models of helicase catalyzed DNA unwinding (31, 32) and this allosteric regulation appears to vary between members of different superfamilies of helicases. For example, the affinity of a monomer of the SF1 E. coli Rep helicase for binding to double-stranded DNA is increased in the presence of AMP-PNP, but decreased in the presence of ADP (32); conversely, the affinity of Rep monomers for binding to single-stranded DNA is decreased in the presence of AMP-PNP, but is not affected by the presence of ADP (32). However, the affinity of the SF2 E. coli RecQ helicase for binding single-stranded DNA is increased by AMP-PNP for a narrow range of nucleotide concentrations, but the affinity for binding either single-stranded or double-stranded DNA is unaffected by the presence of ADP (30). In contrast, the affinity of the SF2 E. coli PriA helicase for binding single-stranded DNA is affected only in the presence of ADP (33). Thus, although the members of the SF1 and SF2 families of helicases have similar structures (34), the allosteric effects of nucleotide binding on their interaction with DNA are different. Based upon the results presented here, we propose that the allosterism between nucleotide and DNA binding in RSC works by a mechanism that is distinct from those which have been previously reported for members of the SF1 and SF2 families of helicases since we observe a decrease in DNA binding affinity in presence of both AMP-PNP and ADP, rather than in the presence of only one nucleotide cofactor.
Implications for DNA translocation and nucleosome repositioning
The ability of RSC to translocate along double-stranded DNA is believed to be central to the mechanism by which the enzyme repositions nucleosomes (12, 13). Interestingly, the processivity of double-stranded DNA translocation by RSC (18, 35), P ~ 0.9, is less than the processivity of single-stranded DNA translocation by either the E. coli Rep (36), P ~ 0.998 , or E. coli UvrD (37, 38), P ~ 0.997, helicases. This may be correlated with the differences in how nucleotide binding by these enzymes allosterically regulates their DNA binding affinity; i.e., the specific sequence at which DNA binding affinity varies during the ATPase cycle, as a result of the allosteric effects of binding ATP, ADP, etc., may influence the processivity of translocation. Specifically, the lower observed processivity for RSC may result from the fact that its motor has a reduced affinity for DNA binding in the presence of both ADP and AMP-PNP, rather than in the presence of only one of these nucleotide cofactors.
The large change in the DNA binding affinity of RSCt between its nucleotide free, ATP-bound, and ADP-bound states suggests a model for DNA translocation by RSCt that is similar to the Brownian ratchet model proposed for single-stranded nucleic acid translocation by the Hepatitis C virus helicase (22, 39). The initial binding of ATP by RSCt, and the associated weakening of the interaction between RSCt and the DNA, would allow for RSCt to move easily along the DNA. Following the hydrolysis of ATP the subsequent RSCt-ADP complex has higher affinity for the DNA and thus becomes fixed in place on the DNA again. This process if finalized by the eventual release of ADP which returns RSCt back to a state with highest DNA affinity. The role of ATP hydrolysis in completion of translocation has also been proposed by Lorch et al (12).
The changes in the pattern of DNaseI digestion of nucleosomal DNA following the binding of RSC to the nucleosome are consistent with the binding of RSC diminishing the interaction of the DNA with the histone octamer (12, 40); additional experiments suggest that both ATP binding and ATP hydrolysis by RSC affect these interactions (12). We have proposed here that the binding of AMP-PNP, an ATP analog, results in a different conformation of the RSCt which has a lower affinity for the DNA. As discussed above, this weakened interaction between the protein and the DNA allows the RSCt to move along the DNA more easily during this phase of the nucleosome repositioning reaction that involves DNA translocation.
ACKNOWLEDGEMENTS
We thank Prof. Mark Richter for the valuable discussions they had with him during the course of this work and also for allowing us to use the fluorescence spectrometer. We would also like to thank Gada Al-Ani, Mike Conner, and Allen Eastlund for discussion throughout the course of this research and for comments on the manuscript.
Abbreviations
- RSC
Remodel the Structure of Chromatin
- RSCt
Remodel the Structure of Chromatin trimer (Used in this study)
- AMP-PNP
Adenylyl-imidodiphosphate
- ADP
Adenosine diphosphate
- NLLS
non-linear least squares
- bp
basepair
Footnotes
This work was supported by National Institutes of Health Grant P20 RR017708 to CJF and NIH GM60415 to BRC. Evan Rich was supported, in part, by an Undergraduate Research Award provided by the Kansas University Honors Program.
REFERENCES
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 3.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 4.Kadonaga JT. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 5.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 6.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 8.Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling. Curr Opin Genet Dev. 2001;11:148–154. doi: 10.1016/s0959-437x(00)00172-6. [DOI] [PubMed] [Google Scholar]
- 9.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 12.Lorch Y, Maier-Davis B, Kornberg RD. Mechanism of chromatin remodeling. Proc Natl Acad Sci U S A. 2010;107:3458–3462. doi: 10.1073/pnas.1000398107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns BR, Peterson CL, Bustamante C. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol Cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cairns BR, Erdjument-Bromage H, Tempst P, Winston F, Kornberg RD. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol Cell. 1998;2:639–651. doi: 10.1016/s1097-2765(00)80162-8. [DOI] [PubMed] [Google Scholar]
- 16.Sirinakis G, Clapier CR, Gao Y, Viswanathan R, Cairns BR, Zhang Y. The RSC chromatin remodelling ATPase translocates DNA with high force and small step size. EMBO J. 30:2364–2372. doi: 10.1038/emboj.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du J, Nasir I, Benton BK, Kladde MP, Laurent BC. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics. 1998;150:987–1005. doi: 10.1093/genetics/150.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer CJ, Saha A, Cairns BR. Kinetic model for the ATP-dependent translocation of Saccharomyces cerevisiae RSC along double-stranded DNA. Biochemistry. 2007;46:12416–12426. doi: 10.1021/bi700930n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams DJ, Hall KB. Monte Carlo applications to thermal and chemical denaturation experiments of nucleic acids and proteins. Methods Enzymol. 2000;321:330–352. doi: 10.1016/s0076-6879(00)21201-9. [DOI] [PubMed] [Google Scholar]
- 20.Straume M, Johnson ML. Monte Carlo method for determining complete confidence probability distributions of estimated model parameters. Methods Enzymol. 1992;210:117–129. doi: 10.1016/0076-6879(92)10009-3. [DOI] [PubMed] [Google Scholar]
- 21.Khaki AR, Field C, Malik S, Niedziela-Majka A, Leavitt SA, Wang R, Hung M, Sakowicz R, Brendza KM, Fischer CJ. The Macroscopic Rate of Nucleic Acid Translocation by Hepatitis C Virus Helicase NS3h Is Dependent on Both Sugar and Base Moieties. J Mol Biol. 2010;400:354–378. doi: 10.1016/j.jmb.2010.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levin MK, Gurjar MM, Patel SS. ATP binding modulates the nucleic acid affinity of hepatitis C virus helicase. J Biol Chem. 2003;278:23311–23316. doi: 10.1074/jbc.M301283200. [DOI] [PubMed] [Google Scholar]
- 23.Lorch Y, Cairns BR, Zhang M, Kornberg RD. Activated RSC-nucleosome complex and persistently altered form of the nucleosome. Cell. 1998;94:29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- 24.Bird LE, Brannigan JA, Subramanya HS, Wigley DB. Characterisation of Bacillus stearothermophilus PcrA helicase: evidence against an active rolling mechanism. Nucleic Acids Res. 1998;26:2686–2693. doi: 10.1093/nar/26.11.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mechanic LE, Latta ME, Matson SW. A region near the C-terminal end of Escherichia coli DNA helicase II is required for single-stranded DNA binding. J Bacteriol. 1999;181:2519–2526. doi: 10.1128/jb.181.8.2519-2526.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong I, Lohman TM. Allosteric effects of nucleotide cofactors on Escherichia coli Rep helicase-DNA binding. Science. 1992;256:350–355. doi: 10.1126/science.256.5055.350. [DOI] [PubMed] [Google Scholar]
- 27.Becker PB. Nucleosome remodelers on track. Nat Struct Mol Biol. 2005;12:732–733. doi: 10.1038/nsmb0905-732. [DOI] [PubMed] [Google Scholar]
- 28.Gorbalenya AE, Koonin EV. Helicases: amino-acid sequence comparisons and structure-function relationships. Current Opinion in Structural Biology. 1993;3:419–429. [Google Scholar]
- 29.Andreeva IE, Roychowdhury A, Szymanski MR, Jezewska MJ, Bujalowski W. Mechanisms of interactions of the nucleotide cofactor with the RepA protein of plasmid RSF1010. Binding dynamics studied using the fluorescence stopped-flow method. Biochemistry. 2009;48:10620–10636. doi: 10.1021/bi900940q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dou SX, Wang PY, Xu HQ, Xi XG. The DNA binding properties of the Escherichia coli RecQ helicase. J Biol Chem. 2004;279:6354–6363. doi: 10.1074/jbc.M311272200. [DOI] [PubMed] [Google Scholar]
- 31.Jezewska MJ, Kim US, Bujalowski W. Interactions of Escherichia coli primary replicative helicase DnaB protein with nucleotide cofactors. Biophys J. 1996;71:2075–2086. doi: 10.1016/S0006-3495(96)79406-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong I, Chao KL, Bujalowski W, Lohman TM. DNA-induced dimerization of the Escherichia coli rep helicase. Allosteric effects of single-stranded and duplex DNA. J Biol Chem. 1992;267:7596–7610. [PubMed] [Google Scholar]
- 33.Lucius AL, Jezewska MJ, Bujalowski W. Allosteric interactions between the nucleotide-binding sites and the ssDNA-binding site in the PriA helicase-ssDNA complex. 3. Biochemistry. 2006;45:7237–7255. doi: 10.1021/bi0518287. [DOI] [PubMed] [Google Scholar]
- 34.Korolev S, Yao N, Lohman TM, Weber PC, Waksman G. Comparisons between the structures of HCV and Rep helicases reveal structural similarities between SF1 and SF2 super-families of helicases. Protein Sci. 1998;7:605–610. doi: 10.1002/pro.5560070309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sirinakis G, Clapier CR, Gao Y, Viswanathan R, Cairns BR, Zhang Y. The RSC chromatin remodelling ATPase translocates DNA with high force and small step size. EMBO J. doi: 10.1038/emboj.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brendza KM, Cheng W, Fischer CJ, Chesnik MA, Niedziela-Majka A, Lohman TM. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc Natl Acad Sci U S A. 2005;102:10076–10081. doi: 10.1073/pnas.0502886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J Mol Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 38.Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levin MK, Gurjar M, Patel SS. A Brownian motor mechanism of translocation and strand separation by hepatitis C virus helicase. Nat Struct Mol Biol. 2005;12:429–435. doi: 10.1038/nsmb920. [DOI] [PubMed] [Google Scholar]
- 40.Chaban Y, Ezeokonkwo C, Chung WH, Zhang F, Kornberg RD, Maier-Davis B, Lorch Y, Asturias FJ. Structure of a RSC-nucleosome complex and insights into chromatin remodeling. Nat Struct Mol Biol. 2008;15:1272–1277. doi: 10.1038/nsmb.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]