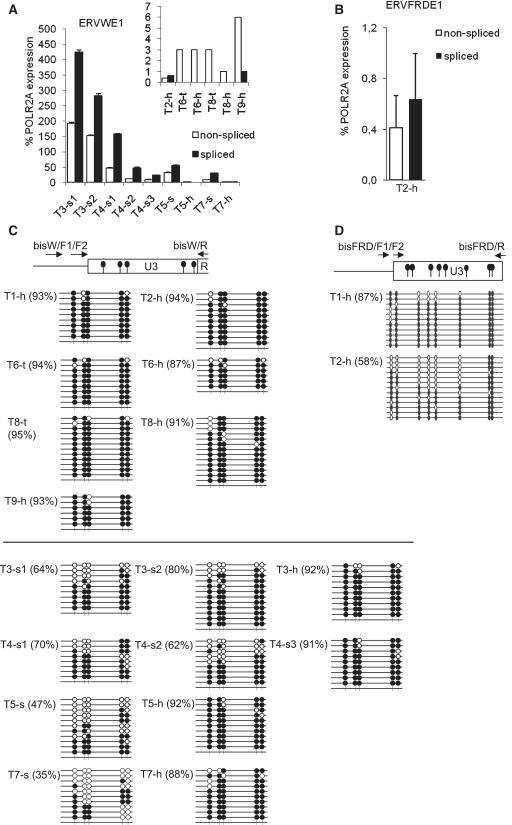
Figure. 3.
Transcription, splicing and CpG methylation of ERVWE1 and ERVFRDE1 in the healthy testes and testicular germ cell tumors. The levels of non-spliced (open columns) and spliced (black columns) transcripts of (A) ERVWE1 and (B) ERVFRDE1 in two samples of healthy testes, T2-h and T9-h, in two samples of granulomatous seminoma 3, T3-s1 and T3-s2, three samples of seminoma 4, T4-s1, T4-s2 and T4-s3, in tumor samples and adjacent healthy tissues of seminoma 5 and 7, T5-s, T5-h, T7-s and T7-h, in tumor sample and adjacent healthy tissue of mixed germ cell tumor 6, T6-t and T6-h and in tumor sample and adjacent healthy tissue of testicular lymphoma 8, T8-t and T8-h were estimated by qRT–PCR and are shown as the average percentage of the POLR2A expression ± SD from three triplicates. The samples of healthy testes and non-seminoma tumors are shown in the inset with different scale of the y-axis. The CpG methylation status of the 5′-LTR U3 region of (C) ERVWE1 and (D) ERVFRDE1 in three samples of healthy testes, T1-h, T2-h and T9-h, and in aforementioned samples of seminomas and non-seminoma tumors was examined by the bisulfite sequencing technique. Methylated CpG sites are indicated by solid circles, unmethylated CpG sites are indicated by open circles, numbers in parentheses depict the percentage of methylated CpG dinucleotides.