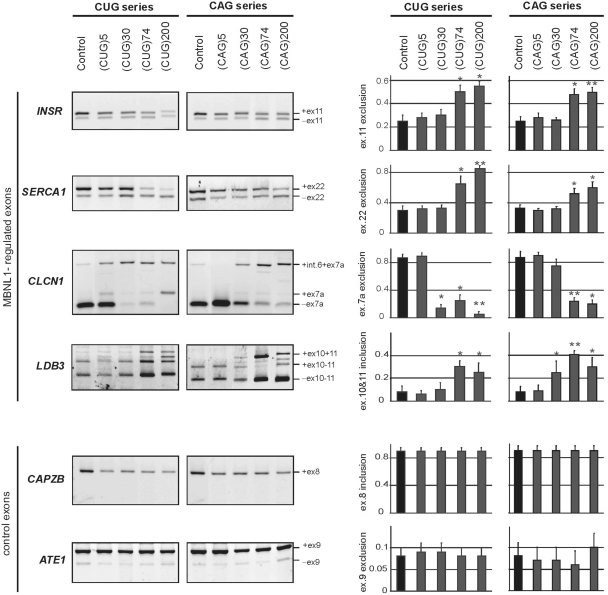
Figure 2.
CUG and CAG repeats trigger similar pattern of the aberrant alternative splicing in HeLa cells. Total RNA isolated from cells of clonal HeLa lines expressing untranslated CUG or CAG repeat RNA of designated lengths was used to assess the levels of splicing variant transcripts from the INSR, SERCA1, CLCN1, LDB3, CAPZB, and ATE1 genes by RT–PCR. DM1-specific splicing aberrations were observed for the longest (74 and 200) both the CUG or CAG repeats and only for genes that are known to be regulated by MBNL1. The CAPZB, that is known to be regulated by CUGBP1 and ATE1 control gene that is not disrupted in DM1 patients, did not show significant changes in the proportion of transcript variants. The experiments were carried out in triplicate and quantitative results are shown as bar diagrams. The fraction of exon inclusion or exclusion (±SD) was calculated by dividing the signal of RT–PCR band corresponding to the inclusion/exclusion splice product by the total RT–PCR representing all splice products. *P = (0.05; 0.001), **P < 0.001 compared with the control.