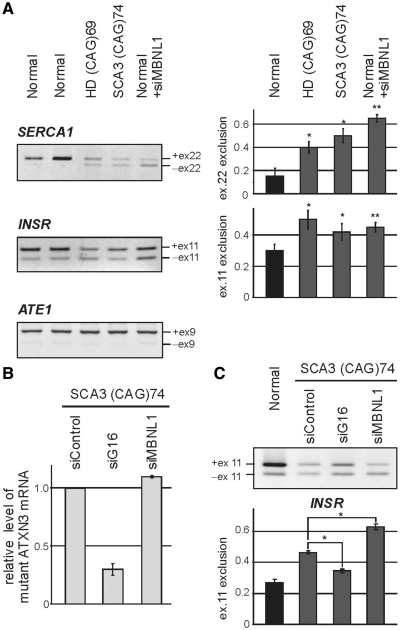
Figure 6.
SERCA1 and INSR undergo alternative splicing changes in human HD and SCA3 fibroblasts. (A) RT–PCR analysis of endogenous SERCA1, INSR and ATE1 mRNA isoforms in fibroblasts derived from HD (69 CAG repeats in HTT) and SCA3 patients (74 CAG repeats in ATXN3) and healthy individuals (Normal). For comparison, normal cells after siRNA-mediated down-regulation of MBNL1 expression were used (+siMBNL1). A modest, but clearly visible disturbance of the balance of the SERCA1 and INSR splicing isoforms was detected in HD and SCA3 cell lines. MBNL1 knock-down resulted in analogical but stronger changes between splicing isoforms. In contrast, ATE1 mRNA expression remained unchanged. The percentages of exon 22 and exon 11 exclusion relative to the total transcripts indicate mean values ± SD of three independent experiments *P = (0.05; 0.001), **P < 0.001 compared with results obtained for normal fibroblasts). (B) The level of mutant ATXN3 transcript (74 CAG repeats) in SCA3 fibroblasts treated with three siRNAs: control, ATXN3-specific (siG16) and MBNL1-specific (siMBNL1). The relative expression level was determined by RT–PCR analysis of CAG repeat region of ATXN3 compared to GAPDH amplification product. (C) Treatment of SCA3 cells with siG16 resulted in partial reversion of INSR splicing misregulation, while MBNL1 silencing led to significant increasing of mRNA isoform without exon 11. Graph shows average values (±SD) for three independent transfection experiments [*P = (0.05; 0.001)].