FIG. 1.
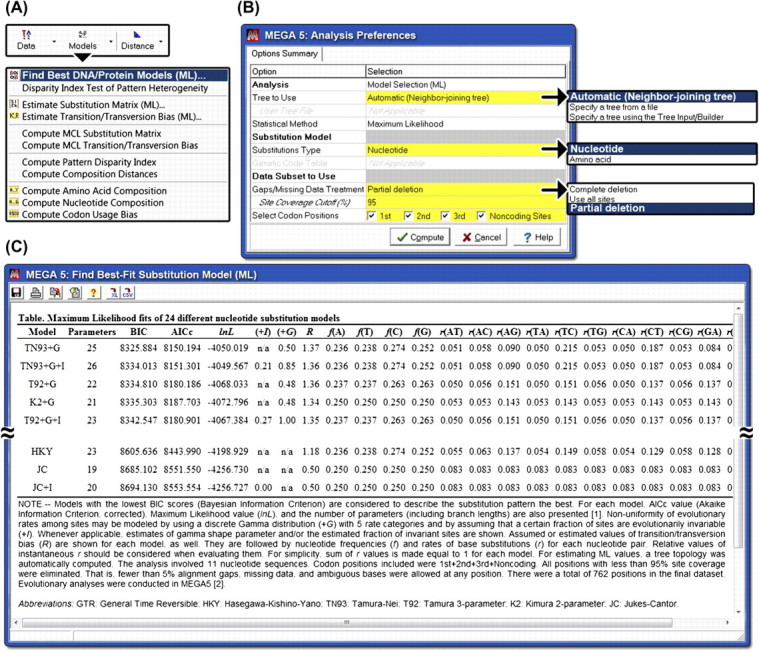
Evaluating the fit of substitution models in MEGA5. (A) The “Models” menu in the “Action Bar” provides access to the facility. (B) An “Analysis Preferences” dialog box provides the user with an array of choices, including the choice of tree to use and the method to treat missing data and alignment gaps. In addition to the “Complete Deletion” and “Pairwise Deletion” options, MEGA5 now includes a “Partial Deletion” option that enables users to exclude positions if they have less than a desired percentage (x%) of site coverage, that is, no more than (100−x)% sequences at a site are allowed to have an alignment gap, missing datum, or ambiguous base/amino acid. For protein coding nucleotide sequences, users can choose to analyze nucleotide or translated amino acid substitutions, with a choice of codon positions in the former. (C) The list of evaluated substitution models along with their relative fits, number of parameters (branch lengths + model parameters), and estimates of evolutionary parameters for Drosophila Adh sequence data which are available in the Examples directory in MEGA5 installation. The note below the table provides a brief description of the results (e.g., ranking of models by BIC), data subset selected, and the analysis option chosen. This figure is available in color online and in black and white in print.
