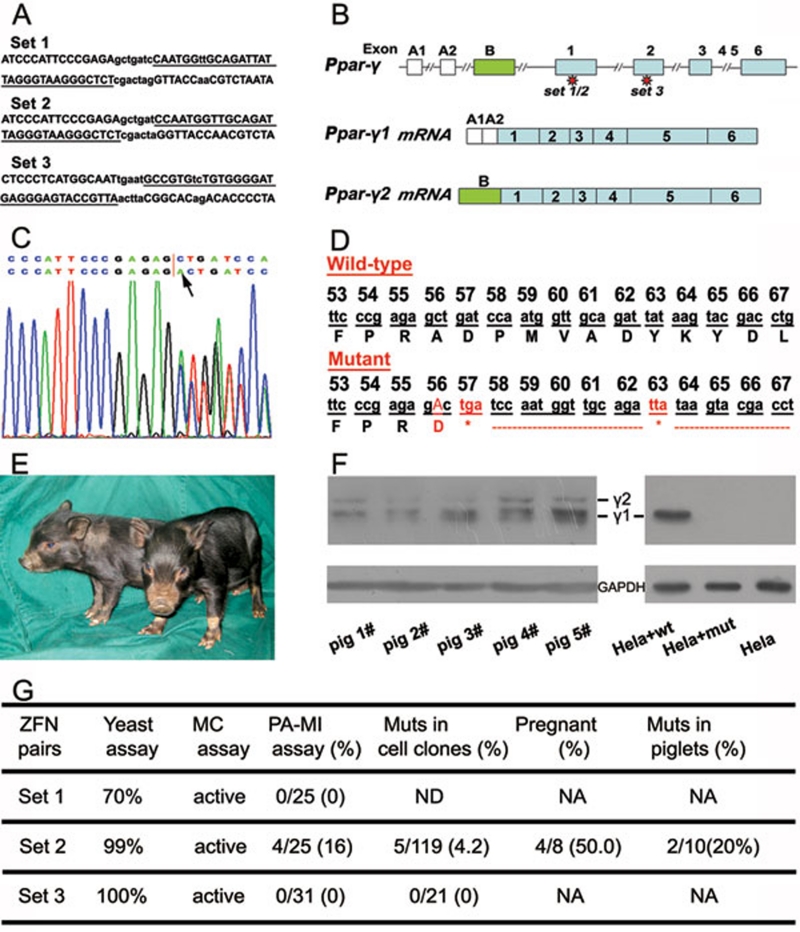
Figure 1.
ZFN-mediated Ppar-γ gene disruption in pigs. (A) Binding sites (underlined) of the zinc-finger nucleases produced. (B) A schematic diagram of the Ppar-γ gene structure. (C) Sequencing diagram of a mutant with a mono-allelic insertion, as indicated by an arrow, at the ZFN-targeting site. The upper line shows the wild-type sequence while the lower line shows the mutant sequence. The diagram shows a double curve after insertion. (D) DNA and amino acid sequences of Ppar-γ proximal to the ZFN target site in cell clone #15 (“*” in red indicates stop codon sites after a frame shift resulted from an insertion at the 56 codon of Ppar-γ1). (E) Piglets confirmed to be Ppar-γ heterozygous knockout. (F) PPAR-γ western blotting. In the left panel, piglets #2 and #3 are mutant piglets while piglets #1, #4 and #5 are wild-type controls. Piglets #1 and #2 are littermates while piglets #3, #4, and #5 are littermates. In the right panel, HeLa cells transfected with wild-type Ppar-γ1 cDNA (HeLa+wt) or with mutated Ppar-γ1 cDNA (HeLa+mut) were used for PPAR-γ western blotting. The untransfected cells were used as the control (HeLa). γ 1 and γ 2 represent the two isoforms of PPAR-γ and GAPDH was used as the internal control. (G) The efficiency of ZFN-mediated targeting in pigs. The yeast assay data were provided by the ZFN manufacturer. ND: not determined; NA: not applicable; MC assay: mammalian cultured cell line-based assay; Muts: Mutants.